Journal of Animal Health and Production
Research Article
Molecular Detection and Characterization of Pasteurella multocida Isolated from Rabbits
Eman H. Mahrous1, Mohamed. W. Abd Al -Azeem2, Faisal A. Wasel1, Waleed Younis2*
1Microbiology Department, Animal Health Research Institute, Sohag Branch, Sohag, B.O. 82525, Egypt; 2Microbiology Department Faculty of Veterinary Medicine South Valley University, Qena, B.O. 83523, Egypt.
Abstract | P. multocida is the main bacterial pathogen causing respiratory manifestations (snuffles) in rabbits with significant economic losses and poor prognosis in Egypt. So, this study aims to detect and identify P. multocida isolated from rabbits by molecular techniques. To establish this goal, lungs, liver, and heart (50 for each organ) were collected from 50 diseased rabbits (suffering from respiratory signs) from backyard rabbits at different localities in Sohag governorate, Egypt. All samples were submitted for PCR test and other conventional methods of identification. Recovery of P. multocida isolates of diseased rabbits from lung, liver, and heart were 23 (46%), 11 (22%), 13 (26%), respectively, with a total incidence of 47 (31.3%). Biochemical reactions proved that 20 (42.5%) out of 47 isolates had the typical biochemical properties of P. multocida. However, only 10 isolates were identified as P. multocida using species-specific primers by conventional PCR with an incidence of 6.7%, besides belonging to serogroup A by multiplex PCR. Serologically only eight isolates belong to somatic serotypes 1, 3, and 12 with a percentage of 20%, 10%, and 10% respectively, while other isolates were untypable. All the recovered isolates were further subjected to multiplex PCR screening of some common virulence genes and revealed the presence of pfhA, hgbB, tbpA, toxA, sodA, ptfA, sodC, nanB, and oma87 with percentages of 30%, 60%, 10%, 100%, 70%, 90%, 90%, 90%, and 100% respectively. Antibiotic susceptibility tests of the recovered isolates revealed that they were all multidrug-resistant (MDR) with a predominance of resistance to erythromycin and oxytetracycline. In conclusion, these findings evidenced that clinically diseased rabbits have a high frequency of MDR and virulent Pasteurella multocida strains.
Keywords | Antimicrobial susceptibility; Pasteurella multocida; Rabbits; Virulence genes
Received | September 15, 2021; Accepted | October 26, 2021; Published | December 13, 2021
*Correspondence | Waleed Younis, Microbiology Department Faculty of Veterinary Medicine South Valley University, Qena, B.O. 83523, Egypt; Email: Wkhrdr39@gmail.com
Citation | Mahrous EH, Abd Al-Azeem MW, Wasel FA, Younis W (2022). Molecular detection and characterization of pasteurella multocida isolated from rabbits. J. Anim. Health Prod. 10(1): 1-9.
DOI | http://dx.doi.org/10.17582/journal.jahp/2022/10.1.1.9
ISSN | 2308-2801
Copyright © 2022 Younis et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Pasteurella multocida is an opportunistic or secondary pathogen that occurs in the respiratory tract of healthy and diseased animals (Dziva et al., 2008). Pasteurella multocida is a Gram-negative, non-spore-forming, non-motile, and capsulated coccobacillus that is penicillin-sensitive and falls in the family pasteurellacae (Kuhnert and Christensen, 2008).
It is a conditional pathogenic microorganism that usually exists in the normal microflora of the mouth, nasopharynx, and upper respiratory tract of mammals, birds, and other species (Wilson and Ho, 2013). Infection with Pasteurella multocida produce many clinical indications in numerous species, including atrophic rhinitis in pigs, fowl cholera in poultry, and hemorrhagic septicemia in buffalo and cattle (Wilson and Ho, 2013). In rabbits, pneumonia, rhinitis, sepsis, mastitis and abscesses are clinical manifestations caused by various strains of Pasteurella multocida (Coudert et al., 2006). Snuffles are a very contagious respiratory disease that causes high deaths in rabbit farms. It can be spread by indirect or direct contact, mainly by aerosol (Premalatha, 2009). Pasteurellosis or snuffles in rabbits are among the most severe diseases that cause significant economic losses in rabbit production units (their use in the laboratory, meat, and fur production) (Stelian et al., 2011).
Serologically, it is classified into five capsular serogroups (A, B, D, E, and F) and 16 lipopolysaccharides (LPS) somatic serotypes (1-16) (Arumugam et al., 2011). Pasteurellosis in rabbits is mainly caused by the capsular type A and, to a lesser extent, type D Pasteurella multocida strains (Dabo et al., 1999). Capsular type F strains have also been recorded in rabbits (Jaglic et al., 2004). Polysaccharide capsules and LPS are some of the main determinants of virulence in the pathogenesis of Pasteurella multocida (Katsuda et al., 2013). However, many other putative virulence factors are related to pathogenicity, including adhesion and colonization factors, fimbriae, iron storage, regulatory proteins, exotoxins, and extracellular enzymes (Katoch et al., 2014).
Using antimicrobial therapy for controlling rabbit pasteurellosis fails to end many infections due to the emergence of multidrug-resistant (MDR) strains with a great potential for transferring this resistance from animals to humans. As a result, the antibiotic resistance phenomenon among pathogenic bacteria has become a growing problem in veterinary and human medical fields (White et al., 2002). This leads to increased treatment costs, prolonged illness, and sometimes death, with adverse effects on the Egyptian economy (Ferreira et al., 2012). Therefore, pre-testing antibiotic susceptibility is essential to select the effective drugs to be used by veterinarians in the field (Varte et al., 2014), as existing data is quite limited. Additionally, the description of co-occurrence between virulence and resistance in Pasteurella multocida from rabbits has not been reported in Egypt. These knowledge gaps encourage the need for researches on these serious issues. Therefore, the present investigation aimed to assess the incidence of Pasteurella multocida in rabbits from different localities in the Sohag governorate as well as their possession of essential virulence-associated genes (ptfA, oma87, hgbB, sodA, sodC, tbpA, pfhA, nanB, and toxA) putatively involved in the pathogenesis of Pasteurella multocida by multiplex PCR.
MATERIAL AND METHODS
Ethics Statement
All animals were handled according to the regulations of the Animal Ethics Committee at the Faculty of Veterinary Medicine, South Valley University (SVU), Qena, Egypt, with good animal practices following the guidelines of the Research Code of Ethics (RCOE-SVU) at the SVU.
Animals and sampling
Fifty diseased rabbits of different ages with respiratory manifestations (oculonasal discharge, sneezing, and coughing) were obtained from different localities in the Sohag governorate. Samples were collected aseptically from (liver, lungs, and heart) organs from each diseased rabbit in a sterile packet and transported to the laboratory in an icebox for bacteriological investigation as soon as possible.
Isolation and identification of P. multocida
A loopful of each sample was inoculated into brain heart infusion broth and incubated for 24 hours at 37 °C. A loopful was taken from incubated broth culture and streaked on blood agar incubated at 37 °C for 24 hours, then subculture on blood agar, MacConkey’s agar, and tryptic soy agar and examined for suspected Pasteurella multocida colonies then subcultured to obtain pure colonies.
Colonies were examined for colonial morphology (shape, size, color, appearance, odor, and elevation). Films were prepared from the suspected pure colonies stained with Gram stain and impression smear directly from samples with Giemsa stain to be examined microscopically under an oil emersion lens (Cruickshank et al., 1975).
Serotyping of Pasteurella multocida by latex agglutination test
The commercial latex agglutination Bonior Mono-kits were used for rapid serological identification of Pasteurella multocida type A according to Sumitha and Rajasekar, (2014).
Molecular characterization of Pasteurella multocida by polymerase chain reaction (PCR)
Confirmation of suspected isolates by conventional PCR: Presumptive isolates were confirmed as Pasteurella multocida by PCR amplification of the species-specific kmT1gene fragment (OIE, 2012). The details of the oligonucleotide primers used to detect species-specific genes are depicted in Table (1).
Identification of capsular type of Pasteurella multocida by multiplex PCR: The capsular types of the confirmed Pasteurella multocida isolates were then determined using the multiplex PCR assay (OIE, 2012). The details of the oligonucleotide primers used to detect capsular genes are depicted in Table (1)
Detection of virulence genes of Pasteurella multocida by multiplex PCR: Pasteurella multocida strains were assessed for the presence of virulence-related genes as filamentous hemagglutinin gene (pfhA), iron acquisition-related factor-B (hgbB), transferrin binding protein (tbpA), dermone
Table 1: Oligonucleotide primers sequences used for identification of Pasteurella species and capsular type.
| Gene function | Target gene |
Primer sequence (5′-3′) |
Amplicon size (bp) | Reference |
|
Pasteurella multocida species specific
|
kmt1 | F- KMT1T7-ATCCGCTATTTACCCAGTGG | 460 | OIE (2012) |
| R- KMT1SP6-GCTGTAAACGAACTCGCCAC | ||||
|
Capsular Type A |
HyaD- | F- TGCCAAAATCGCAGTCAG | 1044 | |
| HyaC | R- TTGCCATCATTGTCAGTG | |||
| B | bcbD | F- CATTTATCCAAGCTCCACC | 760 | |
| R- GCCCGAGAGTTTCAATCC | ||||
| D | dcbF | F- TTACAAAAGAAAGACTAGGAGCCC | 657 | |
| R- CATCTACCCACTCAACCATATCAG | ||||
| E | ecbJ | F- TCCGCAGAAAATTATTGACTC | 511 | |
| R- GCTTGCTGCTTGATTTTGTC | ||||
| F | FcbD | F- AATCGGAGAACGCAGAAATCAG | 851 | |
| R- TTCCGCCGTCAATTACTCTG |
Table 2: Oligonucleotide primers sequences used for detection of virulence genes.
| Target gene |
Oligonucleotide sequence (5′ → 3′) |
Product size (bp) | References |
|
pfhA (F) |
5′ AGCTGATCAAGTGGTGAAC ′3 |
275 |
Cucco et al. (2017)
|
|
pfhA (R) |
5′ TGGTACATTGGTGAATGCTG ′3 |
||
|
hgbB (F) |
5′ACCGCGTTGGAATTATGATTG′3 |
499 |
|
|
hgbB (R) |
5′ATTGAGTACGGCTTGACA′3 | ||
|
tbpA (F) |
5′GAATATTTGGGCGGCAACA′3 |
846 |
|
|
tbpA (R) |
TTCTCGCCCTGTCATCACT′35′ | ||
|
toxA (F) |
5′CTTAGATGAGCGACAAGGT′3 |
846 |
|
|
toxA (R) |
5′GGAATGCCACACCTCT′3 |
||
|
ptfA (F) |
5′TGTGGAATTCAGCATTTTAGTGTGTC′3 |
488 |
Doughty et al. (2000) |
|
ptfA (R) |
5′TCATGAATTCTTATGCGCAAAATCCTGCTGG′3 | ||
|
sodA (F) |
5′TACCAGAATTAGGCTACGC′3 |
361 |
Ewers et al. (2006) |
|
sodA (R) |
5′GAAACGGGTTGCTGCCGCT′3 | ||
|
sodC (F) |
5′AGTTAGTAGCGGGGTTGGCA′3 |
235 |
Ewers et al. (2006)
|
|
sodC (R) |
5′TGGTGCTGGGTGATCATCATG′3 | ||
|
oma87 (F) |
5′ATGAAAAAACTTTTAATTGCGAGC′3 |
948 |
|
|
oma87 (R) |
5′TGACTTGCGCAGTTGCATAAC′3 |
||
|
nanB (F) |
5′GTCCTATAAAGTGACGCCGA′3 |
554 |
|
|
nanB (R) |
5′ACAGCAAAGGAAGACTGTCC′3 |
Table 3: Incidence of Pasteurella multocida isolates by traditional methods.
| Organ | Examined samples | Suspected isolates | No. of positive isolates biochemically | % |
| Lung | 50 | 23 | 11 | 22 % |
| Liver | 50 | 11 | 3 | 6 % |
| Heart | 50 | 13 | 6 | 12 % |
| Total | 150 | 47 | 20 | 13.3% |
crotic toxin (toxA), type IV fimbriae (ptfA), superoxide dismutase (sodA) (Gharibi et al., 2017), superoxide dismutase (sodC), outer membrane protein (oma87), and neuraminidase-B (nanB) genes (Cucco et al., 2017). The details of oligonucleotide primers sequences used to identify virulence genes are depicted in Table (2).
Antibiotic susceptibility testing of Pasteurella multocida
The Kirby–Bauer disk diffusion method was used to examine all the recovered Pasteurella multocida isolates for their antibiotic susceptibility patterns against ten antibiotic agents of four different groups (Bauer et al., 1966). Overnight-grown cultures were spread onto Tryptic Soya Agar (TSA) plates to form a bacterial lawn; after that, antibiotic discs (Oxoid, Hampshire, UK) were evenly placed, and then the plates were incubated at 37 °C for 24 h. The tested antibiotic agents were commonly used in poultry and rabbit’s industry, including ampicillin (AM, 10μg), amoxicillin (AX, 25μg), trimethoprim/sulfamethoxazole (SXT, 25μg), ceftriaxone (CRO,30μg), gentamicin (CN, 10μg), kanamycin (K, 30μg), amikacin (AK, 30μg), erythromycin (E, 15μg), oxytetracycline (T, 30μg), and chloramphenicol (C, 30μg). The inhibition zone diameters were measured and scored as sensitive (S), intermediate (I), and resistant (R) according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2017). Isolates showing resistance to at least three different antibiotic groups were categorized as MDR.
Results
Isolation and identification
The isolation results revealed 23 suspected isolates out of 50 lung samples with a percentage of (46%), while among 50 liver tissue samples, the suspected isolates were 11 with a percentage of (22%), whereas out of 50 heart tissue samples 13 suspected isolates with a percentage of (26%) were revealed. So far, the suspected isolates have been subjected to biochemical identification. The result of biochemical identification revealed that 11 out of 23 suspected Pasteurella isolates from lung tissue showed a typical Pasteurella multocida, 3 out of 11 suspected Pasteurella isolates from liver tissue showed a typical Pasteurella multocida, and 6 out of 13 suspected Pasteurella isolates were confirmed as a typical Pasteurella multocida as shown in Table (3).
Serological identification of Pasteurella isolates from rabbit
The obtained data showed that somatic serotyping of 8 out of 20 tested Pasteurella isolates indicated that 4 (20%) isolates belonged to serotype 1 of Pasteurella multocida, 2 (10%) isolates were serotype 3, and 2 (10%) isolates belonged to serotype 12 while 12 (60%) isolates were untyped as illustrated in Table (4).
Table 4: Serotyping of Pasteurella multocida isolated from rabbits.
| Key No. | Identified bacterium | Serotype |
| 1 | Pasteurella multocida | A: 1 |
| 2 | Pasteurella multocida | A: 3 |
| 3 | Pasteurella multocida | A: 3 |
| 4 | Pasteurella multocida | A: 1 |
| 5 | Pasteurella multocida | A: 1 |
| 6 | Pasteurella multocida | A: 12 |
| 7 | Pasteurella multocida | A: 12 |
| 8 | Pasteurella multocida | A: 1 |
Identification of Pasteurella isolates by conventional PCR:
The obtained data showed that 20 Pasteurella isolates were submitted for identification of kmT1 gene by PCR and found that 10 (50%) out of all isolates (5 from lung tissue, 3 from liver tissue, and 2 from heart tissue) were positive to kmT1 gene and confirmed to be Pasteurella multocida strains as shown in Figure (1).
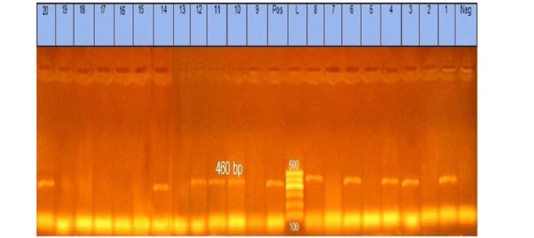
Figure 1: Agarose gel showing Pasteurella multocida PCR amplified product of 460 bp for kmT1 gene using specific primer. Lane L: marker, 100bp DNA ladder. Lane 1, 3, 4, 6, 8, 10, 11, 12, 14, and 20: strains belong to Pasteurella multocida isolates
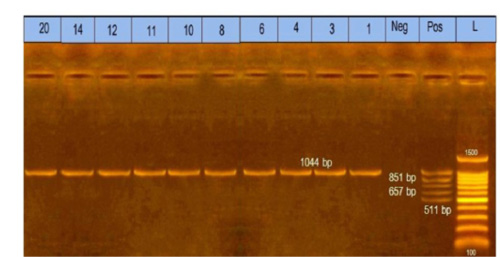
Figure 2: Multiplex polymerase chain reaction for capsular typing. Lane L: marker, 100bp DNA ladder. Lane1, 3, 4, 6, 8, 10, 11, 12, 14, and 20: strains belong to capsular A type.
Table 5: Antimicrobial sensitivity testing of Pasteurella multocida isolated from rabbits in Sohag governorate.
| Therapeutic agent | Symbol | Concentration (mcg) |
Resistant No. (%) |
Sensitivity No. (%) |
|
Trimethoprim/sulfamethoxazole |
SXT | 25 | 0 | 10 (100%) |
|
Gentamicin |
CN | 10 | 1 (10%) | 9 (90%) |
|
Amoxicillin |
AX | 25 | 7 (70%) | 3 (30%) |
|
Ceftriaxone |
CRO | 30 | 8 (80%) | 2 (20%) |
|
Amikacin |
AK | 30 | 4 (40%) |
6 (60%) |
| Kanamycin | K | 30 | 8 (80%) | 2 (20%) |
|
Ampicillin |
AM | 10 | 6 (60%) | 4(40%) |
|
Chloramphenicol |
C | 30 | 5 (50%) | 5(50%) |
|
Oxytetracycline |
T | 30 | 10 (100%) | 0 |
|
Erythromycin |
E | 15 | 10 (100%) |
0 |
Capsular type of Pasteurella multocida isolates by multiplex PCR
The obtained data showed that 10 Pasteurella isolates were submitted to identify capsular type by multiplex PCR and found that 10 (100%) isolates belonged to capsular type A, as shown in Figure (2).
Incidence of virulence genes of Pasteurella multocida in rabbit by multiplex PCR
These results showed that the recovered isolates were further subjected to multiplex PCR screening for some common virulence genes and revealed the presence of pfhA, hgbB, tbpA, toxA, sodA, ptfA, sodC, nanB, and oma87 with a percentage of 30%, 60%, 10%, 10%, 70%, 90%, 90%, 90%, and 100%, respectively, as shown in Figures (3-5).
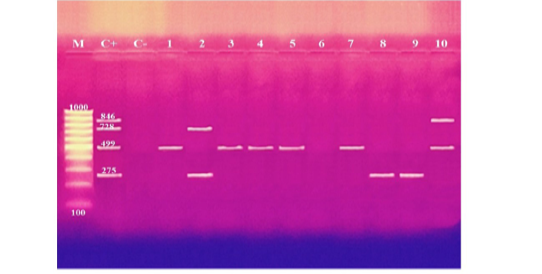
Figure 3: Agarose gel electrophoresis of multiplex PCR of pfhA (275 bp), hgbB (499 bp), tbpA (728 bp), and toxA (846 bp) virulence genes for characterization of Pasteurella multocida. Lane M: 100 bp ladder as a molecular size DNA marker. Lane C+: Control positive for pfhA, hgbB, tbpA and toxA genes. Lane C-: Control negative. Lanes 1, 3, 4, 5, 7 & 10: Positive strains for hgbB gene. Lanes 2, 8 & 9: Positive strains for pfhA gene. Lane 2: Positive strain for tbpA gene. Lane 10: Positive strain for toxA gene. Lane 6: Negative strain for pfhA, hgbB, tbpA and toxA genes.
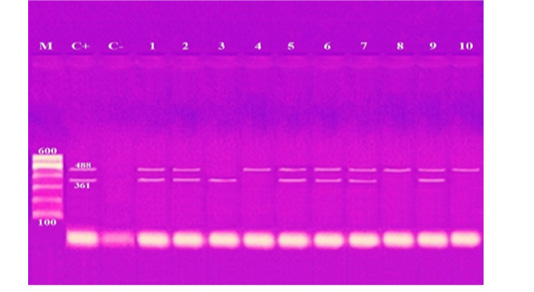
Figure 4: Agarose gel electrophoresis of multiplex PCR of sodA (361 bp) and ptfA (488 bp) virulence genes for characterization of Pasteurella multocida. Lane M: 100 bp ladder as a molecular size DNA marker. Lane C+: Control positive for ptfA and sodA genes. Lane C-: Control negative. Lanes 1, 2, 5, 6, 7 & 9: Positive strains for ptfA and sodA genes. Lanes 4, 8 & 10: Positive strain for ptfA gene. Lanes 3: Positive strain for sodA gene
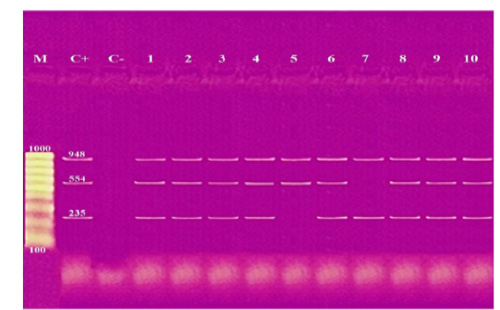
Figure 5: Agarose gel electrophoresis of multiplex PCR of sodC (235 bp), nanB (544 bp), and oma87 (948 bp) virulence genes for characterization of Pasteurella multocida. Lane M: 100 bp ladder as a molecular size DNA marker. Lane C+: Control positive for sodC, nanB, and oma87 genes. Lane C-: Control negative. Lanes 1, 2, 3, 4, 6, 8, 9 & 10: Positive strains for sodC, nanB, and oma87 genes. Lane 5: Positive strain for nanB and oma87 genes. Lane 7: Positive strain for sodC and oma87 genes.
Antimicrobial sensitivity testing of Pasteurella multocida isolates
The obtained data showed that antibiotic susceptibility testing of the recovered isolates revealed that they were all multidrug-resistant (MDR) with a predominance resistance to erythromycin and oxytetracycline (100% for each), followed by kanamycin and ceftriaxone (80% for each). In contrast, they were sensitive to trimethoprim/sulfamethoxazole, gentamycin, amoxicillin, amikacin, ampicillin, and chloramphenicol, as shown in Table (5).
Discussion
In Egypt, Pasteurella multocida is a common pathogen of rabbits, causing severe epidemics and significant economic losses in the rabbit breeding industry (Nasser et al., 2013). All isolates obtained in this study were subjected to complete phenotypic properties. The data revealed that all isolates were tiny white colonies on tryptic soy agar. While in 5% of sheep blood agar appears as a non-hemolytic small yellowish-white colony (dew drop-like colony approximately 1 mm in diameter). Films were prepared from the suspected pure colonies stained with Gram showed Gram-negative, non-spore-forming, non-motile, and coccobacilli shaped bacteria with impression smear directly from samples stained with Giemsa stain showed bipolarity under an oil immersion lens. These results agree with (Mohamed et al., 2020; Ehsan, 2019) who reported the same results. Biochemical reactions of isolates revealed that all isolates were positive for oxidase, catalase, and indole test but negative for urea hydrolysis. Also, all isolates were able to ferment glucose, mannitol and could not ferment lactose. These results go hand in hand with the previous results of Tinella et al. (2020) and Awad and Abd El-Hamid, (2019).
The incidence of Pasteurella multocida from diseased rabbits using traditional methods were 20 (13.3%) isolates, as shown in Table (3). These results nearly coincided with those obtained by Ehsan, (2019) who found that 15 (10%) suspected isolates from a pure culture were morphologically and biochemically identified, but these results seemed to be lower than Stelian et al. (2011) with an incidence of (77.55%). The difference in distribution frequency may be due to the individuality of immunological status and health of the sampled rabbits and/or environmental conditions (Stelian et al., 2011). High temperature, poor nutrition, and transportation may increase the incidence of Pasteurella multocida and vice versa. Also, the variation may be due to the method of detection used or the locality in which the study was done. The recorded data in Table (4), illustrated that serologically 8 isolates belong to somatic serotypes 1, 3, and 12 with a percentage of 4 (20 %), 2 (10%), and 2 (10%), respectively, by using latex agglutination test. These results nearly coincide with those obtained by Mahrous, (2017) and Sumitha and Rajasekar, (2014). The obtained results in this study revealed that 10 out of 20 isolates were positive for Pasteurella multocida with a percentage of 50% of the examined isolates. The specificity of the primer was confirmed by positive amplification of the fragment with the extracted DNA of the bacterial isolates at the expected size of 460 bp, as shown in Figure (1). These results are nearly in accordance with Mohamed et al. (2020) and Awad and Abd El-Hamid, (2019).
The obtained results in this study revealed that all ten isolates of Pasteurella multocida of diseased rabbits belonged to capsular type A. The specificity of the primer was confirmed by positive amplification of the fragment with the extracted DNA of the bacterial isolates at the expected size of 1044bp, as shown in Figure (2). These results nearly coincided with that reported by previous workers (Krishna et al., 2017; Ehsan, 2019). The obtained results of this study proved that the incidence of Pasteurella multocida isolates from diseased rabbits by PCR was (6.7%) of total samples. These results nearly coincided with that reported by (Mahrous, 2017) who found that the incidence of Pasteurella multocida isolated from diseased and apparently healthy rabbits by PCR was (11.6%). However, these results seemed higher than those reported by Ehsan, (2019), who found that the incidence of Pasteurella multocida isolated from diseased and apparently healthy rabbits by PCR was 2%.
In this study, Pasteurella multocida isolates were screened for the existence of nine critical virulence-associated genes (ptfA, omP87, hgbB, sodA, sodC, tbpA, pfhA, nanB, and toxA) involved in bacterial pathogenesis (Shirzad and Tabatabaei, 2016). The obtained results in this study showed that the virulence-associated genes were found among all the isolates, as shown in Figures (3-5). Our results were confirmed by other researchers (Sarangi et al., 2014; Ferreira et al., 2012), who reported the occurrence of selected virulence genes encoding (dermonecrotoxin production (toxA), fimbriae adhesions (ptfA), extracellular enzymes (nanB), outer membrane protein synthesis (omA87), filamentous hemagglutinin (pfhA), superoxide dismutase (sodA), superoxide dismutase (sodC), transferrin binding protein (tbpA), and hemoglobin-binding protein (hgbB)) in Pasteurella multocida isolated from rabbits.
The high prevalence rate (90%) of the ptfA gene detected in this study was expected because it is assumed to be a critical factor in fixing the bacteria on the epithelial cells’ surfaces. These results nearly coincided with that reported by Ferreira et al. (2012). Moreover, the nanB gene was found in most examined isolates (90%), as previously evidenced by Sarangi et al. (2014). This could be attributed to the fact that sialidases (nanB) play an important role in colonization to the host’s epithelial surfaces and promote the virulence of bacteria by unmasking the key receptors and reducing mucin effectiveness. Concerning the toxA gene, only one isolate harbored this gene. The negative results of all specimen for the toxA gene may be associated to the fact that the gene encoding dermonecrotic toxin is more common in atrophic rhinitis of swine. This gene was not common in poultry and sheep strains (Ewers et al., 2006). It has been found that even in toxin-producing pig isolates, the presence of this gene is low, and the gene is usually lost after several subcultures. Some researchers described that the toxA gene is not inserted into the bacterial chromosome but inserted into the lysogenic phage that infects the pathogen (Pullinger et al., 2004). Gene pfhA was also found in the three strains analyzed, which contrasts with that reported by Ferreira et al. (2012), who reported that gene pfhA was not observed in any tested strain. Also, as stated to date, the presence of this gene in Pasteurella multocida strains from rabbits was not detected in any research study. The tbpA gene is closely allied to ruminant strains (buffalo, cattle, and sheep), which clarifies their little occurrence in the present study (10%) (Atashpaz et al., 2009; Ewers et al., 2006).
Genes encoding proteins with different functions, such as hemoglobin-binding proteins (hgbB), superoxide dismutase (sodA, sodC), and membrane proteins (oma87), showed frequencies like those reported by Ewers et al. (2006) and Tang et al. (2009).
The antibiotic susceptibility test of the isolates showed that they all showed multidrug resistance (MDR) with a predominance of resistance to erythromycin and oxytetracycline (100% for each), followed by kanamycin (90% for each). These results were like those reported by Awad and Abd El-Hamid, (2019). On the other hand, the data on the antibiotic susceptibility test of rabbit Pasteurella multocida isolates, available in another literature conducted in Italy, showed a relatively low frequency of resistance against tetracycline (Cucco et al., 2017). Different geographic locations of the isolated strains might have caused these variations (Awad and Abd El-Hamid, 2019).
Concerning the resistance profiles in our study, the MDR phenomenon was observed among all recovered isolates. This observation parallels a recent report conducted in Egypt (El-sayed et al., 2018), where all the examined rabbit Pasteurella multocida strains demonstrated MDR patterns remarkably. The presence of multi-resistant Pasteurella multocida isolates recovered from rabbits affected by pasteurellosis has already been reported in Italy (Cucco et al., 2017). In another study in Brazil, (47.8%) (22/46) of rabbit Pasteurella multocida strains were resistant to at least one of the tested drugs (Ferreira et al., 2012). Antibiotic resistance is a growing problem probably attributed to the extensive and indiscriminate usage of antibiotics to prevent and treat pasteurellosis in the field.
CONCLUSION
Pasteurella multocida is one of the most severe pathogens in rabbits; it causes significant economic losses to rabbit farms. Molecular characterization of Pasteurella multocida by PCR analysis is a more sensitive and specific method for detecting and identifying Pasteurella multocida than conventional rapid biochemical and serological tests. In clinically diseased rabbits, a high frequency of MDR and virulent Pasteurella multocida strains were observed. Our observations highlight the role of rabbits as potential sources of Pasteurella multocida with a wide range of virulence determinants and multidrug-resistant isolates with subsequent significant adverse public health implications.
Conflict of interest
The authors declare that there is no conflict of interest for this publication.
Authors contributions
All authors contributed equally to study design, sampling, methodology, interpretation of results, and manuscript writing.
REFERENCES





