Journal of Infection and Molecular Biology
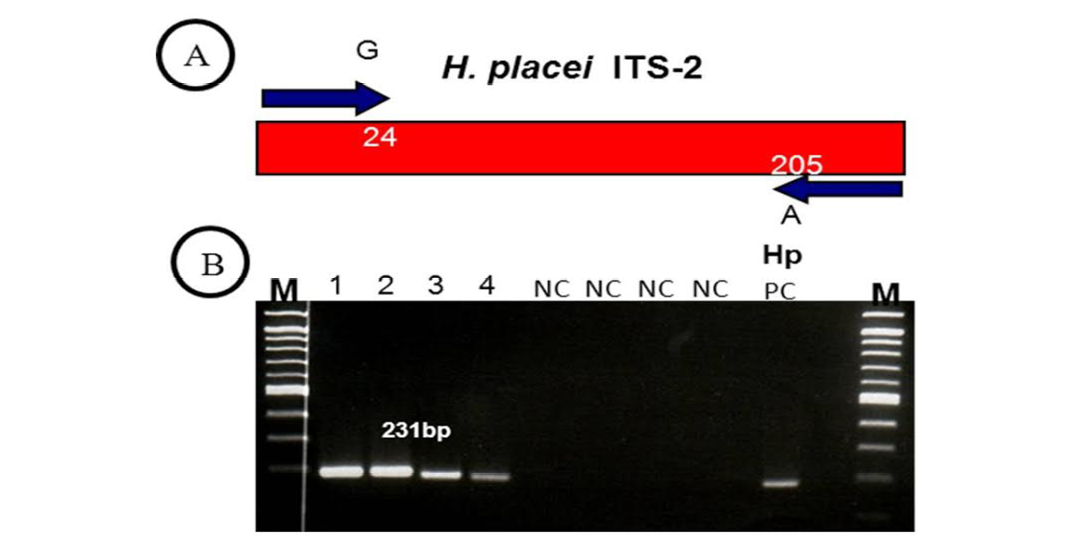
(A) H. placei has fixed nucleotide sequence differences at position 24 and 205 of the ITS-2 sequence. This allowed the design of forward and reverse primers for which the final 3’ base was complementary to these positions and so specific respective species. (B) ITS-2 products amplified with H. placei allele-specific primers. M, 100bp ladder; Tracks 1-4; amplicons from H. placei single worm DNA templates, Tracks 5-8; (NC) no template negative control; HP PC, H. placei positive control
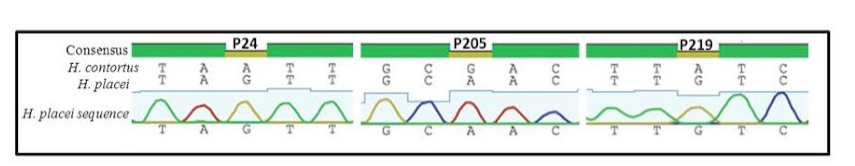
Sequence chromatogram of 1 out of 66 sequences (named H. placei sequences) obtained from 4 populations showing three fixed species-specific positions (P24, P205, P219) of the rDNA ITS-2 sequence of H. placei