Journal of Infection and Molecular Biology
Short Communication
Genetic Variation in the rDNA ITS-2 Sequence of Haemonchus placei from Cattle Host
Qasim Ali1, Muhammad Imran Rashid1, Kamran Ashraf1, Muhammad Nauman Zahid2, Shoaib Ashraf 4, Umer Chaudhry 3
1Department of Parasitology, University of Veterinary and Animal Science, Lahore, Pakistan; 2 Department of Microbiology, University of Veterinary and Animal Science, Lahore, Pakistan; 3 Department of Comparative Biology and Experimental Medicine, Faculty of Veterinary Medicine, University of Calgary, Alberta, Canada; 4 Department of Pharmacology, University of Veterinary and Animal Science, Lahore, Pakistan.
Abstract | The large stomach worm, Haemonchus, commonly known as the barber’s pole worm, is a blood sucking nematode found in the abomasa of small and large ruminants. Allele-specific amplification of the rDNA internal transcribed spacer-2 (ITS-2) sequences was performed from the total of 78 individual adult worms to screen Haemonchus placei at species level, which is the significant diagnostic tool to identify this major economically important species. Further full sequences analysis of the ITS-2 region revealed that there are 4 sites shows intraspecific variations at position 65, 111, 125 and 148. For instance this study is the first documented report of intraspecific genetic variations in the rDNA ITS-2 sequences of H. placei from cattle in Pakistan and the results shows that H. placei is genetically different from the isolates studied previously. However, detailed and large size samples strategy will be required to identify the co-infection and interspecies hybridization between H. placei and Haemonchus contortus in cattle.
Keywords | Haemonchus placei, rDNA ITS-2, Pakistan
Editor | Tahir Yaqub, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | August 20, 2014; Revised | October 13, 2014; Accepted | October 25, 2014; Published | December 02, 2014
*Correspondence | Umer Chaudhry, University of Calgary, Alberta, Canada; Email: unchaudh@ucalgary.ca
Citation | Ali Q, Rashid MI, Ashraf K, Zahid MN, Ashraf S, Chaudhry U (2015). Genetic variation in the rDNA ITS-2 sequence of Haemonchus placei from cattle host. J. Inf. Mol. Biol. 3(1): 13-18.
DOI | http://dx.doi.org/10.14737/journal.jimb/2015/3.1.13.18
ISSN (Online) | 2307-5465; ISSN (Print) | 2307-5716
Copyright © 2015 Ali et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Gastrointestinal parasitic (GI) nematodes are extremely important agents of both human and animal disease (Sutherland and Leathwick, 2011). GI nematode infections constitute a significant threat to the health and welfare of grazing livestock throughout the world and impose a significant cost in terms of productivity in grazing animals. GI parasitic infections of livestock are among the most economically important diseases in the livestock of Pakistan (Annonymous, 2012-2013).
The major GI nematode species in cattle are H. placei, Ostertagia ostertagi, Mecistocirrus digitatus and Trichostrongylus axei, Strongyloides papillosus, Cooperia and Nematodirus and Oesophagostomum radiatum found in the abomasum, small intestine and large intestine (Oku et al., 1987; Fukumoto et al., 1990). Of these species, H. placei, O. ostertagi and Cooperia oncophora are considered the most prevalent in cattle; in particular, the infection rate of H. placei was high. This nematode is commonly described as a large stomach worm, namely, a trichostrongyloid nematode, and is an important blood-sucking nematode present in the abomasum of cattle. This parasite may cause mucosal inflammation, haemorrhage, ulcers and necrosis in the abomasum (Gennari et al., 1991). It appears to be confined mainly to Asian countries; however, it has also been found in North America, Australia, and Brazil. The movement of its hosts for agricultural purposes has resulted in the global spread of this parasite.
Generally, ruminants are concurrently infected with more than one species of GI nematodes, each having a different pathological effect on the host. It has been a difficult task to eradicate GI nematodes from grazing ruminants due to the variation in host susceptibility to the parasite, the wide distribution of nematodes in nature and the presence of wild ruminants. An effective method is required for the control of nematodes by reducing their infection rate and their transmission to the host in order to protect cattle for production losses. Strategies for the control of nematodes by using anthelmintic drugs should be devised based on the quantitation and identification of species (Mochizuki et al., 2006).
The rDNA ITS-2 sequences are the most commonly used markers to discriminate among nematode species (Gasser and Newton, 2000). This sequence has been a popular choice because of its variability and is under concerted evolution (Nadler et al., 2000). Thus the ITS-2 gene has been widely applied in species identification within the genus Haemonchus (Gasser, 1999; Heise et al., 1999). For example, the separate species status of H. placei and H. contortus was supported by ITS-2 data, where three fixed nucleotide differences at position 24, 205 and 219 were reported by Stevenson et al. (1995) and Chaudhry et al. (2014). There have only been a few documented studies on the genetic variation within the isolates of H. placei, however the variations within H. contortus ranged between 4.0 to 5.2% (Heise et al., 1999; Stevenson et al., 1995 and Chaudhry et al., 2014). Genetic characterization is important for accurate identification and effective control due to the anthelmintic resistance problem in this nematode (Gasser, 2006). The present study contributes to validate the allele-specific molecular base marker and genetic variations in the ITS-2 region of this important nematode species from cattle host.
Adult worms were harvested on necropsy from the abomasa of ruminant hosts collected from abattoirs in Punjab province of Pakistan (4 populations) (Table 1). Following ethanol fixation, worms were examined under a dissecting microscope to determine whether they belongs to Haemonchus genera based on size and gross appearance (Lichtenfels and Pilitt, 2000). Species identity was subsequently confirmed by molecular methods as described below. Overall, the dataset was composed of 78 individual specimens of adult worms from the genus Haemonchus distributed among four different populations from individual cattle host.
Adult worms were fixed in 70% ethanol immediately following removal from the host abomasum. The heads of individual worms were dissected and lysed in single 0.2ul tube containing 50µl of proteinase K lysis buffer and stored at -80°C as previously described (Redman et al., 2008). A neat single worm lysate dilution, 1µl of 1:5, was used as PCR template and identical dilutions of lysate buffer, made in parallel, were used as negative controls.
The rDNA ITS-2 region was amplified from individual Haemonchus adult worm lysates using a allele-specific forward primer complementary to 5’ prim end of rDNA ITS-2 coding sequence (HpITS-2F: 5’-atactacaatgtggctag-3’) and reverse primer complimentary to the 3’ prim end of rDNA coding sequence (HpITS-2R: 5’- TGATAAAAGAACATCGTT-3’). A 231bp fragment spanning the entire ITS-2 rDNA region was PCR amplified using a 50µl master mix containing final concentrations of 1X thermopol reaction buffer, 2mM MgSO4, 100uM of each dNTPs, 0.1uM forward and reverse primers and 1.25U Taq DNA polymerase (New England Biolabs). Thermo-cycling parameters were 95oC for 5 min followed by 35 cycles of 95oC for 1 min, 55oC for 1 min and 72oC for 1 min with a single final extension cycle of 72oC for 5 min.
The entire rDNA ITS-2 region was amplified from Haemonchus (identified as a H. placei section 3.1) adult worm lysates using a “universal” forward primer complementary to 5.8S rDNA coding sequence (NC1F: 5’-acg tct ggt tca ggg ttg tt- 3’) and reverse primer complimentary to the 28S rDNA coding sequence (NC2R: 5’-TTA GTT TCT TTT CCT CCG CT- 3’) (Stevenson et al., 1995). A 321bp fragment spanning the entire ITS-2 rDNA region was PCR amplified using a 50µl master mix containing final concentrations of 1X thermopol reaction buffer, 2mM MgSO4, 100uM of each dNTPs, 0.1uM Positions 24, 205 and 219 (indicated in larger font) have fixed interspecies polymorphisms whereas the other listed positions show intra-specific variation. rDNA ITS-2 sequence polymorphisms identified from a total of 66 individual derived from four H. placei populations: H1C, H2C, H3C, and H4C forward and reverse primers and 1.25U Taq DNA polymerase (New England Biolabs). Thermo-cycling parameters were 95oC for 5 min followed by 35 cycles of 95oC for 1 min, 57oC for 1 min and 72oC for 1 min with a single final extension cycle of 72oC for 5 min. rDNA ITS-2 amplicons from individual worm lysates were further were sequenced using the reverse primer (NC2R: 5-TTA GTT TCT TTT CCT CCG CT- 3’) on an ABI Prism 377 capillary sequencer. Sequences were edited and aligned with H. placei ITS-2 sequences available in Genebank (Acc No X78812) using Geneious Pro 5.4 software (http://www.geneious.com/).
Table 1: Summary of the field populations of Haemonchus collected from cattle host from Punjab (PN) region of Pakistan
|
Field populations ID |
Host |
Worm number |
Origin |
District |
|
H1C |
Cattle |
33 |
Abattoir |
Sargodha |
|
H2C |
Cattle |
23 |
Abattoir |
Lahore |
|
H3C |
Cattle |
7 |
Abattoir |
Okara |
|
H4C |
Cattle |
16 |
Abattoir |
Sahiwal |
Table 2: Summary of inter- and intra-species variation in the H. placei rDNA ITS-2 sequence.
|
Species |
Alignment position |
Nucleotide |
Type of base exchange |
|
H. placei populations |
24 |
G |
Fix SNP |
|
65 |
A/T |
Transversion |
|
|
111 |
A/G |
Transition |
|
|
125 |
C/T |
Transition |
|
|
148 |
A/T |
Transversion |
|
|
205 |
A |
Fix SNP |
|
|
219 |
G |
Fix SNP |
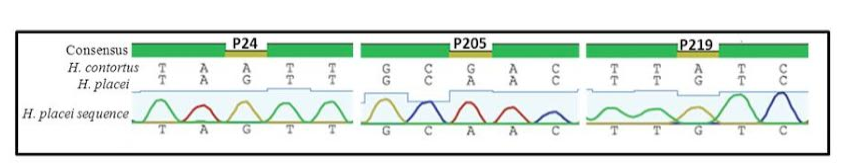
Figure 1: Sequence chromatogram of 1 out of 66 sequences (named H. placei sequences) obtained from 4 populations showing three fixed species-specific positions (P24, P205, P219) of the rDNA ITS-2 sequence of H. placei
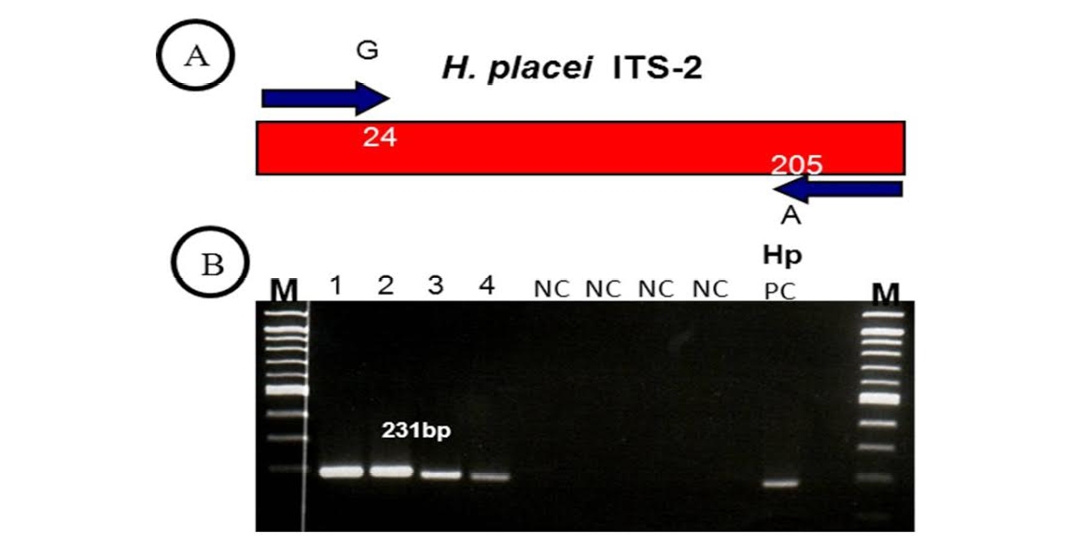
Figure 2: (A, B)
(A) H. placei has fixed nucleotide sequence differences at position 24 and 205 of the ITS-2 sequence. This allowed the design of forward and reverse primers for which the final 3’ base was complementary to these positions and so specific respective species. (B) ITS-2 products amplified with H. placei allele-specific primers. M, 100bp ladder; Tracks 1-4; amplicons from H. placei single worm DNA templates, Tracks 5-8; (NC) no template negative control; HP PC, H. placei positive control.
Previous work has identified three single nucleotide polymorphisms (SNPs) in the rDNA ITS-2 (positions 24, 205 and 219) that show fixed sequence specific differences between H. placei and other closely related species H. contortus (Stevenson et al., 1995). However this study was based on just two H. placei and eight H. contortus worms obtained from cattle and sheep respectively. Consequently, Chaudhry et al. (2014) further provided that these inter-species differences were fixed when larger numbers of individuals from geographically diverse populations were examined. In the present study, allele-specific PCR primers were developed based on P24 and P219 SNPs, which will then be used to identify Haemonchus populations for the presence of H. placei. For this purpose, allele-specific PCR was then applied to screen all 78 individual worms (from 4 populations) collected from cattle and a total of 66 worms were identified H. placei, rest of the 12 worms did not amplified with this assay (Figure 1).
The rDNA ITS-2 sequence was PCR amplified and sequenced from DNA derived from 66 worms in four Haemonchus populations (identified H. placei in section 3.1). In all cases, H. placei sequences contained P24 (G), P205 (A), P219 (G) confirming SNPs (Table 2 and Figure 2) previously identified by Stevenson et al. (1995) and Chaudhry et al. (2014). In addition to confirming the three species-specific fixed SNPs, there were 4 sites that showed intraspecific variation in H. placei at position 65,111, 125 and 148 (Table 2).
Accurate identification of parasite species is crucial not only for diagnosis, treatment and control but epidemiological studies and clinical trials. Although, sufficient morphological differences exist among adult and larval GI parasitic nematodes to allow their accurate identification, the availability of similar techniques for nematode eggs remains an obstacle to reliable diagnosis. In some cases the structure and size of the egg can be diagnostic; however, in many instances, similarities among eggs from different species and even distinct genera require alternative methods for their differentiation. Presently, the method commonly utilized for GI parasitic nematodes involves in-vitro cultivation of egg to infective third stage larval (L3) recovery and direct collection of adult parasite from slaughtered animals followed by morphological identification. These procedures are labour intensive, time consuming and prone to errors due to variation in egg viability and parasite development in culture. The advent of DNA technology has provided alternative approaches for the identification of parasites and molecular techniques like allele-specific PCR have proven to be useful in species identification. Studies have shown that the rDNA ITS-2 contain reliable genetic markers to distinguish closely-related species of trichostrongylid nematodes (Gasser et al., 1993; Campbell et al., 1995; Chilton et al., 1995; Stevenson et al., 1995; Wimmer et al., 2004).
Haemonchus spp. are important, highly pathogenic blood feeding parasites of cattle in many parts of the world, particular in warmer regions with high humidity, such as Asia, south America and the southern USA. H. placei is the most common species traditionally reported from cattle (Gasbarre et al., 2009a; Gasbarre et al., 2009b). Of the four populations examined (one from Sargodha, one from Lahore, one from Okara and one from Sahiwal), the prevalence of 85% H. placei suggesting this is still the predominant Haemonchus species infecting cattle in Pakistan.
Indeed, inter-species variation in the ITS-2 sequences between H. placei and H. contortus in cattle, sheep and goats host revealed minor differences such as substitutions of three residues at position P24, P205 and P219, many investigators also indicate the intra-specific variation of ITS-2 sequences among H. contortus from domestic ruminant in the world, however very limited information about the genetic variability in H. placei in ruminants currently available worldwide and particularly on the south Asian region. This work investigates further insight into genetic relationships of H. placei from Pakistani region. Therefore we se quenced ITS-2 region of 66 H. placei individuals and our analysis was conducted from four different cattle host to avoid strain specific nucleotide variations. The results demonstrate three species specific SNPs [P24 (G), P205 (A), P219 (G)] and four intra-specific variations sites at position 65,111, 125 and 148 in H. placei (Table 2). Of those, position 65, 123 and 148 have been previously shown in H. placei (Stevenson et al., 1995; Chaudhry et al., 2014). However position 111 polymorphic site had not been identified previously, so we hypothesized that P111 is a new intra-specific variation present in H. placei from Pakistani region. The newly identified genetic variation in the ITS-2 sequence represents a significant addition to databases.
To our knowledge, this is the first study in Pakistan exploring the genetic variation in H. placei. These results can further use to monitor Haemonchus infection in a context of gastro-intestinal control approaches further integrated to a sustainable agriculture. In or der to confirm the development of H. placei resistant to anthelmintic in cattle further study must be carried out.
References




