Journal of Infection and Molecular Biology
Research Article
Characterization and Transcriptional Profile of Toll-Like Receptors of Bubalus bubalis with Endometritis
Noha Mohamed Osman1,2*, Ahlam Ahmed Abou Mossallam2, Fawzy Reyad El Seedy3, Eman Roshdy Mahfouz2, Rui Li4, Michal Mikula5
1Department of Molecular and Computational Biology, University of Southern California, 1050 Childs Way RRI 316 Los Angeles, CA 90089, USA; 2Department of Cell Biology, Genetic Engineering Division, National Research Center, Dokki, Giza, Egypt; 3Department of Bacteriology, Mycology and Immunology, Faculty of Veterinary Medicine, Beni-Suef University, Beni-Suef, Egypt; 4Department of Animal Sciences, Washington State University, Pullman, WA 99164-6351, USA; 5Department of Oncological Genetics, Maria Sklodowska-Curie Memorial Cancer Center and Institute of Oncology, Warsaw, Poland.
Abstract | Toll-like receptors (TLRs) are a vital component of innate immune system and perform an important role in disease resistance through recognition of pathogen associated molecular patterns (PAMPs) and initiation of a convenient immune response. Endometritis is the most important disease causing infertility in cattle and buffalo and TLRs play a critical role in elimination of that disease. The present study aims to characterize the transcripts of TLRs: TLR1, TLR2, TLR4, TLR6 and TLR10 in Bubalus bubalis with endometritis. Quantitative real-time (q-PCR) assays were performed to detect transcript expression profiles of these TLRs in liver, mammary gland, ovary and uterus of Bubalus bubalis with and without endometritis. The result showed that the transcript profiles of TLRs especially TLR2 and TLR4 were significantly differences in mammary glands, ovary and uterus but on the othe hands, there is no significant difference in the transcript profile of TLRs in liver with or without endometritis. TLRs nucleotide sequences have more than 97% nucleotide homology between Bubalus bubalis with and without endometritis. Simple modular architecture tool (SMART) analysis revealed the presence of TIR domains and different numbers of leucine rich repeat motifs in tested TLRs. In conclusion, for animals infected with endometritis, there was a significant difference of TLR2 and TLR4 in uterine tissue compared to uninfected animals. Therefore, it is suggested to consider the TLRs results as detection markers for endometritis.
Keywords | Toll-Like receptors (TLRs), Bubalus bubalis, Endometritis, Gene expression
Editor | Tahir Yaqub, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | January 11, 2015; Revised | February 17, 2015; Accepted | February 19, 2015; Published | March 03, 2015
*Correspondence | Noha Mohamed Osman, University of Southern California, Los Angeles, USA; Email: nmo_138@usc.edu, Osman.noha@yahoo.com
Citation | Osman NM, Mossallam AAA, El Seedy FR, Mahfouz ER, Li R, Mikula R (2015). Characterization and transcriptional profile of toll-like receptors of Bubalus bubalis with endometritis. J. Inf. Mol. Biol. 3(2): 34-46.
DOI | http://dx.doi.org/10.14737/journal.jimb/2015/3.2.34.46
ISSN (Online) | 2307-5465; ISSN (Print) | 2307-5716
Copyright © 2015 Osman et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
The innate immune system is a global system of host defense against pathogens. Recognition of pathogens relied on a set of germ line-encoded receptors recognized pattern-recognition receptors (PRRs) as a part of innate immune recognition. These receptors have the ability to recognize conserved pathogen-associated molecular patterns (PAMPs) produced by large groups of pathogens (Akira, 2001; Janeway and Medzhitov, 2002).
Ten members of the TLR receptor family are broadly expressed in the mammalian genome (Akira et al., 2006). TLR1, TLR2 and TLR6 recognize bacterial lipids such as lipoteichoic acid (LTA), whereas TLR3, TLR7, and TLR8 as well as TLR9 recognize nucleic acids, often from viruses. TLR9 also identifies bacterial DNA. TLR4 recognizes lipopolysaccharide (LPS) from Gram-negative bacteria such as E. coli, while TLR5 binds to flagellin. The ligand for TLR10 is still not known (Hirata et al., 2007; Regan et al., 2013).
TLRs are type 1 transmembrane proteins with N terminal extracellular leucine-rich repeat (LRR) and C terminal intracellular TIR (Toll/IL 1 receptor) domain. A TIR domain is required for initiating intracellular signaling and inducing downstream effector molecules, while LRRs are responsible for the recognition of PAMPs. (Akira et al., 2006; Kumar et al., 2009; Dubey et al., 2013).
TLRs are expressed broadly in Antigen-presenting cells, and regulate the activation of both innate and adaptive immunity through recognition of specific patterns of microbial components (Medzhitov, 2001; Matsushima et al., 2007). Their expression and distribution are associated with altered immune responsiveness (Helmby and Grencis, 2003).
TLRs play an important role in regulation of ovulation, fertilization, gestation and parturition, as well as in pathological conditions such as endometritis and mastitis (Kannaki et al., 2011). The endometrium is the first line of defense against microbial pathogens. The endometrial cells have ability to develop innate immune defense in cattle, humans and rodents (Herath et al., 2006; Davies et al., 2008). Endometritis is the most common uterine disease observed in buffaloes slaughtered at abattoirs, and is one of the main causes of infertility in the cattle and buffalo (Azawi et al., 2008; Ajevar et al., 2014).
Microbial disease of the female genital tract is most common, and has greatest economic importance in both humans and animals. Microbial infections of the genital tract cause infertility by disrupting uterine and ovarian function (O’Neill, 2008). Establishment of infection in the endometrium occurs by the interaction between the virulence factors of microbial invaders and the endometrial immune defenses (Sheldon et al., 2010).
Bubalus bubalis population (195 million heads) includes both river and swamp buffaloes. Egyptian buffaloes are of the river type with a population of 4 million heads; it ranks the third after Indian (115 millions) and Pakistani (31.7 millions) buffalo, which plays an important role in the economy of developing countries (FAOstat, 2013). The present study was aimed to characterize the TLRs genes and their transcriptional profile in Bubalus bubalis with and without endometritis.
Materials and Methods
Tissue Samples
Tissue samples from four different organs; liver, ovary, mammary gland and uterus, were obtained from thirty animals with and without endometritis from a local abattoir immediately after slaughter and kept on ice until processing. The uteri of animals with endometritis had abnormal secretions as profuse purulent exudate, with signs of inflammation as swelling, redness and hardness (Williams et al., 2005; Sheldon et al., 2006). These animals had no evidence of other diseases based on visual inspection and culture of bacteria using standard microbiological techniques.
RNA Isolation and Reverse Transcription
Total RNA was extracted from liver, ovary, mammary gland and uterus using Easy –RED™ kit (iNtRON Biotechnology) according to the manufacturers’ protocols. The RNA quality and purity was determined by NanoDrop 1000 (Thermo Scientific) and cDNA was synthesized as per manufacturer’s procedure (RevertAid First Strand Synthesis Kit, Thermo Scientific).
Primer Design and Optimization of RT-PCR
The TLRs coding regions were amplified by designing primers using Bubalus bubalis transcripts sequences as following: TLR1 (HQ327989), TLR2 (HM756162), TLR4 (HQ343416), TLR6 (HQ327992), TLR10 (HQ327991) as presented in Table 1 using the Primer3 software (Rozen and Skaletsky, 2000). These primers were used for PCR analysis and Real time quantitative PCR (qRT-PCR). The primers were synthesized by Amersham Pharmacia Biotech Company.
Table 1: Primers sequences of TLR1, 2, 4, 6 and 10 used in PCR
|
Gene |
Name |
Forward primer (5’-3’) |
Reverseprimer (5’-3’) |
Annealing |
Size(bp) |
|
TLR1 |
TLR1.1 TLR1.2 TLR1.3 |
ATG CCT GAC ATC CTC TCA CT TGA CCC AGG AAA TGA AAT CT TCA ACC AGG AAC TGG AAT ACT TGG A |
ACC TCC TTG GAT CGA GGT CC CAG CAG AAA TAC ATT AAC ACG G TGG GCA GGG CAT CAA ATG GA |
63.9°C 63.9°C 60°C |
1165 bp 1195 bp 120 bp |
|
TLR2 |
TLR2.1 TLR2.2 TLR2.3 |
AGA GTG GAG GTC AAA TCA CTG GAC A GCC TGT AAG GAT GCC TGG CCC GGT TTT AAG GCA GAA TCG TTT G |
TGC CTG AAA CTT GTC GGT GGC C GGT CAG TCT TGG TCT GGT GGT AAG GCA CTG GGT TAA ACT GTG T |
68.6°C 68.6°C 60°C |
1329 bp 1253 bp 190 bp |
|
TLR4 |
TLR4.1 TLR4.2 TLR4.3 |
CGG CAC AGA CAG AGG GTC ATG C TTT TGG GAC AAC CAA CCT GAA GCA T CTT GCG TAC AGG TTG TTC CTA A |
GCA AAC CTT GAA AAT GGC AGG CAA CAA GAA GTG ACA ACC TCC ACC TGA CAT AAC TTC TCC AGC TTC CCA G |
62.2°C 63.6°C 60°C |
1500 bp 1342 bp 153 bp |
|
TLR6 |
TLR6.1 TLR6.2 TLR6.3 |
ATC AGC AGC AAC CCT CCG GG ACA CAT GCT TTG TCC TCA GGA ACC A AAG TGA ATG ATG ACA ATT GTC |
TGG GTG TGG TAT CTT TAG CAG CCT
AAT TTA TCC CCT GCT GCG TTT GGA
AGT GAC TTC AAT GTC GTT T |
67°C 65°C 60°C |
1532 bp 1388 bp 235 bp |
|
TLR10 |
TLR10.1 TLR10.2 TLR10.3 |
TCT GTG GAG CCG CTG CAA GC GCC AGC AAC ACA TCC TTG AGG C GCT GCA AGC CTG AGG GAT TT |
TGG ACT TTT TCC AGA GCT GCC AGG GCC TGT AGA GCC CCA CCC ACT AGG ATT TTA TCA CTG GTT CAC CTC |
62°C 54°C 60°C |
1654 bp 1270 bp 93 bp |
|
RPL0 |
RPL0 |
CAA CCC TGA AGT GCT TGA CAT |
AGG CAG ATG GAT CAG CCA |
60°C |
220 bp |
|
GAPDH |
GAPDH |
CCT GGA GAA ACC TGC CAA GT |
GCC GTA TTC ATT GTC ATA CCA |
60°C |
214 bp |
For qPCR analysis, large ribosomal protein (RPL0) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) used as an internal calibrator due to their threshold cycle (CT) values are close to those of TLRs. Each PCR reaction was performed using 2 µl of 50-60 ng cDNA template, 12.5 μl of RNAse DNAse free water, 2.5μl of 2mM dNTPs, 2.5 μl of 10X taq polymerase buffer, and 2.5 μl forward and reverse primers (10 µM), as well as 0.25µl (1U) of Taq polymerase in 25 µl reaction final volume. The following cycling conditions were used: 3 min. at 94ºC; 35 cycles for 1 min at 94ºC; 2 min at specific annealing of each primer as in table (1); 1 min at 72ºC and a final extension for 10 min at 72ºC. Following optimization, the presence of a single product was confirmed on a 1.5% agarose gel by electrophoresis. The gels were inspected by UV light and photographed by Gel documentation system (In Genius, Syngenebioimaging).
The PCR products were purified using (Thermo Scientific, USA) purification kit, according to the manufacturer’s instructions. The purified product was sequenced using ABI PRISM Dye Terminator Cycle Sequencing Kit (Applied Biosystems) in an automatic sequencer (ABI Prism3100, Genetic analyzer (Applied Biosystems).
Sequence Analysis
cDNAs infected uterus were used to amplify TLR1, TLR2, TLR4, TLR6 and TLR10 genes. Then, the amplified PCR products were sequenced using reverse and forward primers.
Sequence alignment was carried out using NCBI/BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) between coding regions (CDs) and amino acids of tested TLRs in infected uterus of Bubalus bubalis with endometritis and Bubalus bubalis sequences in GenBank database.
Characterization of the TLRs Domains
The different functional domains of TLRs (The transmembrane regions (TM); Leucine-rich repeats (LRR); LRR C-terminal (LRR-CT) and Toll/IL-1 receptor (TIR) was predicted by using simple modular architecture analysis tool (SMART), available at http://smart.embl-heidelberg.de/ (Schultz et al., 1998) and LRRfinder (www.lrrfinder.com) (Offord et al., 2010). The amino acid sequence of TIR domains of tested TLRs of infected animals with endometritis was subjected to align with TIR amino acid sequence of Bubalus bubalis sequences in GenBank database for detecting the percentage of the identity.
Quantitative Real Time PCR (qRT-PCR)
qRT-PCR was used to estimate TLR expression levels in the examined samples. For each sample, cDNA was diluted 1:25 with 10Mm Tris pH 7.5. For each sample, 1.9 µl of diluted cDNA template was combined with 2 µl of 2X Master Mix (Bioline) and 0.1 µl of 10 µM primer set for a final reaction volume 4 µl in MicroAmp optical 384 well reaction plate in triplicates along with no template control (NTC) to all wells. Cycling was performed on 7900HT Real-Time PCR system (Applied Biosystems, Life Technologies Corporation) using the following cycling conditions: 95°C for 10 min (hold), 95°C for 15 sec (melt) and 60°C for 1 min (anneal/extend) for 40 cycles. Dissociation curves were generated to ensure that a single amplicon had been produced.
Statistical Analysis
The expression of candidate genes was evaluated using a 2-ΔΔCt method (Livak and Schmittgen 2001) .The Geometric mean of reference genes (RPLP0 and GAPDH) acted as an internal calibrator within each sample to calculate Δ Ct.The ΔΔ Ct was calculated by ΔCt (treatment) − ΔCt (control). The fold change in expression was then used as a relative measure of gene expression. Data analysis was carried out by non-parametric Mann-Whitney test. Significance was set at P<0.05 and the results were reported as mean ± SEM.
Results
Sequencing of TLRs genes
The full-length of TLR1, TLR2, TLR4, TLR6, and TLR10 cDNAs coding regions of Bubalus bubalis with endometritis have been submitted to GenBank and their accession numbers are presented in Table 2. The percentage of nucleotide and aminoacid identity of Bubalus bubalis with and without endometritis is shown in Table 2. There was a highly degree of identity between Bubalus bubalis with and without endometritis.
Analysis of Functional Domains of Tested TLRs
The results showed that all TLRs of Bubalus bubalis with and without endometritis have LRR and TIR domains and one transmembrane domains (TM) in all tested TLRs except TLR6, which has two transmembrane domains (TM). LRRCT and LRRNT (cysteine flanking regions of LRR at C and N terminals, respectively) were examined in tested TLRs, we found all TLRs contain LRRCT, while LRRNT was absent in all TLRs as shown in Figure 1.
Next, we used LRR finder to predict the repeat number of LRR motifs present in TLRs. As shown in Table 3, we found an equal number of LRRs (20) in TLR2, TLR6 and TLR10 Bubalus bubalis with and without endometritis but on the other hand TLR1 of Bubalus bubalis with and without endometritis has (18) and (19) LRR respectively, while TLR4 of Bubalus bubalis with and without endometritis has (21) and (22) LRRs with 2 LRRs predicted overlapped respectively.
Table 2: Coding DNA, translated amino acids and TIR domain sequence similarity of TLRs between Bubalus bubalis with and without endometritis
|
TLR |
CDs identity (%) |
Amino acid identity (%) |
TIR domain identity(%) |
Accession number |
|
TLR1 |
99.5 |
99.3 |
100 |
KM058706 |
|
TLR2 |
99.6 |
99.1 |
99.3 |
KM058707 |
|
TLR4 |
99.5 |
98.9 |
99.3 |
KM058708 |
|
TLR6 |
99.8 |
99.2 |
98.6 |
KM058709 |
|
TLR10 |
99.5 |
98.9 |
97.9 |
KM058710 |
Table 3: Leucine-rich repeats in amino acid sequences of tested TLRs
Species |
TLR1 |
TLR2 |
TLR4 |
TLR6 |
TLR10 |
Bubalus bubalis with endometritis |
18 |
20 |
21 and 2 predicted overlapped |
20 |
20 |
Bubalus bubalis |
19 |
20 |
22 and 2 predicted overlapped |
20 |
20 |
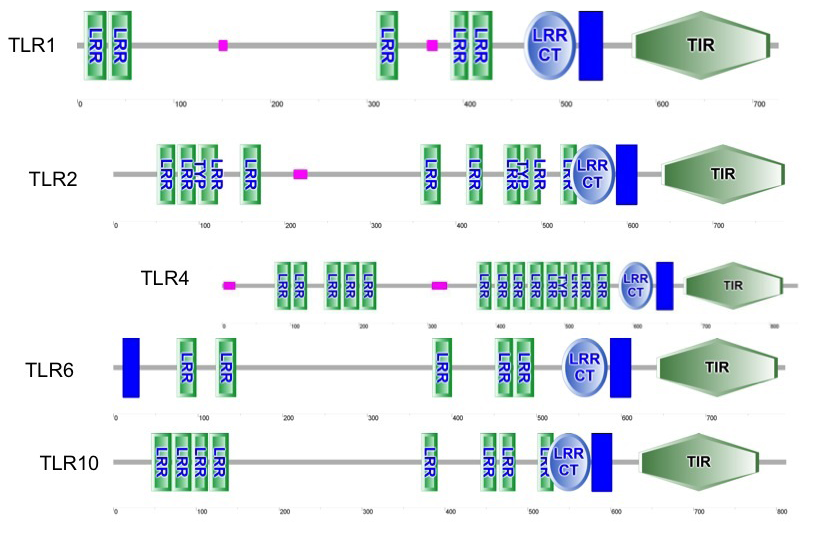
Figure 1: predicted functional domains in tested TLRs of Bubalus bubalis, with and without endometritis (vertical bar is transmembrane domain (TM)
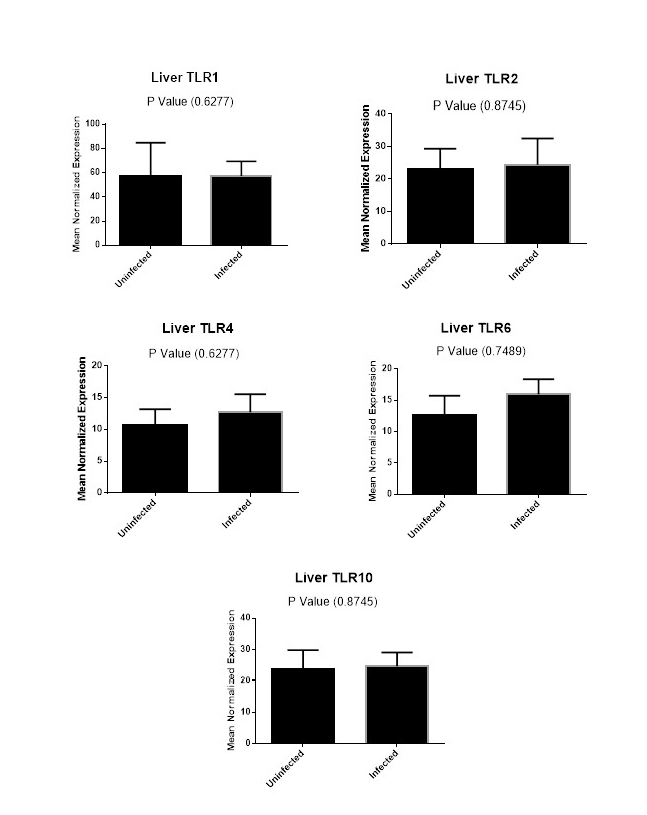
Figure (2A): Expression levels of TLR1,TLR2, TLR4,TLR6 and TLR10 genes in liver tissues collected from animals with and without endometritis. The expression levels of these genes were normalized to RPL0 and GAPDH and calibrated with the average of control. Expression levels were obtained from three pooled biological replicate samples ± S.E.M.
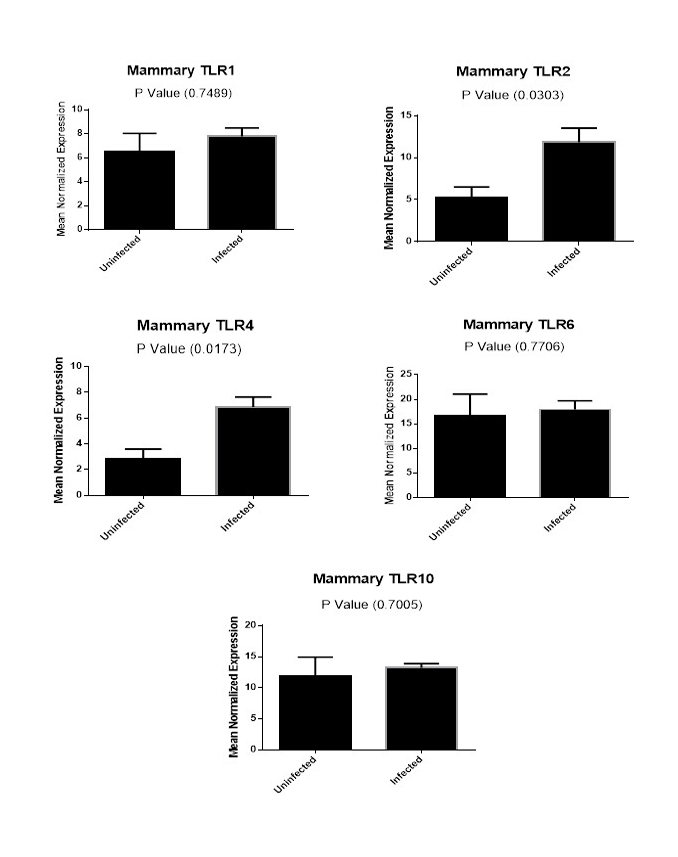
Figure (2B): Expression levels of TLR1,TLR2, TLR4,TLR6 and TLR10 genes in mammary gland tissues collected from animals with and without endometritis. The expression levels of these genes were normalized to RPL0 and GAPDH and calibrated with the average of control. Expression levels were obtained from three pooled biological replicate samples ± S.E.M.
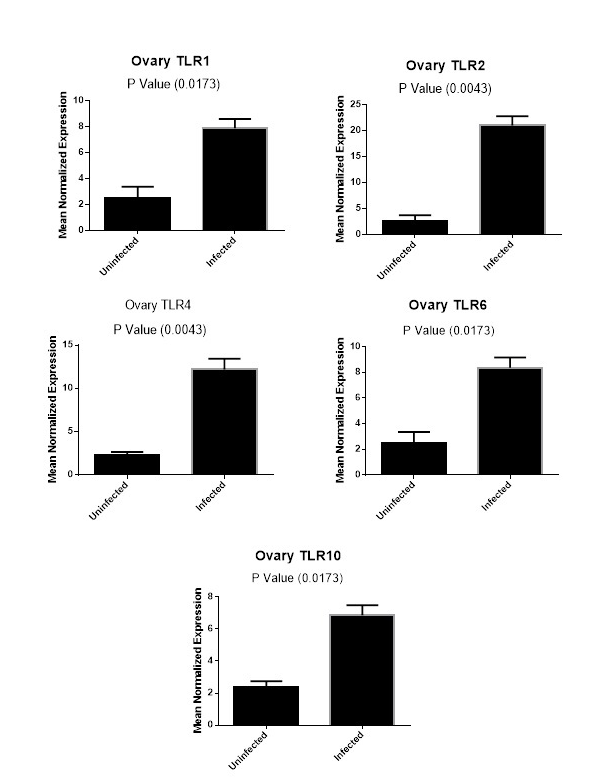
Figure (2C): Expression levels of TLR1,TLR2, TLR4,TLR6 and TLR10 genes in ovarian tissues collected from animals with and without endometritis. The expression levels of these genes were normalized to RPL0 and GAPDH and calibrated with the average of control. Expression levels were obtained from three pooled biological replicate samples ± S.E.M.
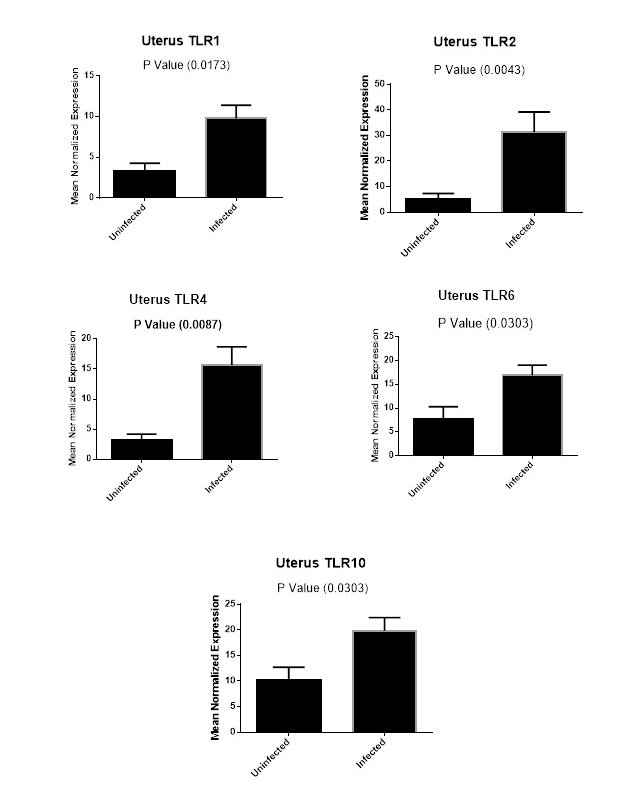
Figure (2D): Expression levels of TLR1,TLR2, TLR4,TLR6 and TLR10 genes in uterine tissues collected from animals with and without endometritis. The expression levels of these genes were normalized to RPL0 and GAPDH and calibrated with the average of control. Expression levels were obtained from three pooled biological replicate samples ± S.E.M.
Results of Expression of TLRs Genes
As shown in Figure 2D, our results revealed that TLR1 expression levels increased six-fold in the uterine tissues of Bubalus bubalis with endometritis compared to those in the uterine tissues of animals without endometritis (P=0.0173). Similarly, the expression levels of both TLR6 and TLR10 were higher with nine-fold increase for TLR6 (P=0.0303), and an almost ten-fold increase for TLR10 (P=0.0303) in the uterine tissues of animals with endometritis compared to those in the uterine tissues of animals without endometritis. In addition, TLR2 showed twenty-six-fold increase in the uterine tissues of animals with endometritis compared to those in the uterine tissues of animals without endometritis (P=0.0043). Moreover, TLR4 showed a significant increase in the uterine tissues of animals with endometritis compared to those in the uterine tissues of animals without endometritis, with a twelve-fold increase (P= 0.0087).
In the ovarian tissues, the expression profile of candidate genes showed that the expression of TLR1, TLR6 and TLR10 had the same level of expression between animals with and without endometritis (P =0.0173, Figure 2C). In addition, there was a significant increase in the expression levels of TLR2 and TLR4 genes in the ovary tissues of animals with endometritis compared with those without endometritis (P=0.0043; Figure 2C).
A comparison of the expression levels of TLR1, TLR2, TLR4, TLR6 and TLR10 genes in mammary gland tissues from animals with and without endometritis were performed. As shown in Figure 2B, the expression levels of TLR1 (P=0.7489), TLR6 (P=0.7706) and TLR10 (P=0.7005) genes in the mammary gland tissues had no significant difference between animals with and without endometritis. Conversely, TLR2 showed a highly significant increase in animals with endometritis compared to animals without endometritis, with a 6.6 fold increase (P=0.0303). Similarly, TLR4 showed a highly significant increase in the mammary glands of animals with endometritis compared to those from animals without endometritis, with a 4.0 fold increase (P=0.0173).
Finally, we carried out a comparison for the expression levels of TLR1, TLR2, TLR4, TLR6 and TLR10 genes in liver tissues from animals with and without endometritis. As shown in Figure 2A, these tissues had similar expression levels for TLR1, TLR2, TLR4, TLR6 and TLR10.
Discussion
In the present study, we reported the first sequencing data of TLRs genes and their expression in Bubalus bubalis with endometritis. Toll-like receptors consider the initial defence against microbes in endometrium. Their characterizations are important to evaluate the innate immune system. TLR1, TLR2, TLR4, TLR6 and TLR10 cDNA, translated amino acids and TIR domain sequences of Bubalus bubalis with endometritis were compared with Bubalus bubalis without endometritis. TLRs cDNA ncleotide sequences of Bubalus bubalis with and without endometritis were 98 to 99% identical. Furthermore, TLR6 of Bubalus bubalis with and without endometritis had an insertion of nucleotide triple ‘TTA’ resulting in the addition of tyrosine amino acid (Y) at position 181 compared with Bos Taurus (Carl et al., 2003; Dubey et al., 2013).
TIR domain has an important role in activation of inflammatory response, in cooperation with the adaptor protein MyD88 (Kawai and Akira, 2007). We found that the identity of TIR sequences between with and without endometritis ranged from 97 (TLR10) to 99.3 (TLR2 and TLR4) and 100 %( TLR1). TLR2 recognizes peptidoglycan cell wall components of Gram-positive bacteria, while TLR4 ligands include Gram-negative bacteria. Both of TLR2 and TLR4 have an important role in regulation of innate immune response (Herath et al., 2006; Davies et al., 2008).
The number and structure of LRRs of TLRs have a significant effect on the function of pathogen recognition receptor system among species (Kumar et al., 2009; Dubey et al., 2013). Peter et al. (2009) reported that deletion of LRR motif leads to loss of functional integrity of the ligand–binding domain of murine TLRs. In addition, (Dubey et al., 2013) demonstrated that the differences in the number of LRRs among buffalo and cattle may lead to possible differences in the specific recognition of TLR ligands between these species. In our study we found that the number of LRR domains of Bubalus bubalis with endometritis and their distribution is similar to Bubalus bubalis without endometritis except TLR1 and TLR4.
Bacterial infection of the female genital tract is common in animals especially after parturition causing infertility and sometimes mortality (Davies et al., 2008). The endometrium is the first defence against pathogens. PAMPs were recognized by TLRs which consider as key components of innate immune system which lead to increase expression of TLRs (Akira, 2006). TLRs expression were evaluated in liver, mammary gland, ovary and uterus of Bubalus bubalis with and without endometritis. The results showed no significant difference in TLRs expression in liver tissues between buffaloes with and without endometritis but on the other hand the expression of TLRs in mammary gland showed no significant difference in the expression levels of TLR1, TLR6 and TLR10 between animals with and without endometritis. Conversely, TLR2 and TLR4 genes expression significantly increased in mammary glands of animals with endometritis compared to animals without endometritis. In Ovarian tissue, TLR1, TLR6 and TLR10 had the same level of significant difference (P=0.0173). Similarly, TLR2 and TLR4 had the same level of significant difference (P=0.0043). This indicates that the expression levels of several TLRs vary significantly in association with the degree of infection in animal (Sharma et al., 2006; Deborah et al., 2008; Chapwanya et al., 2009; Priyanka et al., 2012). The presence of uterine infection due to pathogenic infection had a localized effect on ovarian function (Sheldon et al., 2002). This is hypothesis is supported by (Williams et al., 2008) who stated that pathogenic agents such as E.coli has the ability to produce endotoxin lipopolysaccharide (LPS), which is released from infected uterine tissue to ovarian tissue and peripheral circulation leading to disruption of the hypothalamus,pituitary and ovary functions in animals.
Previous studies demonstrated that the expression levels of TLRs family members increase upon endometritis. Specifically, the mRNA levels of TLR2 and TLR4 increase in endometritis infection of bacterial origin (Ozinsky et al., 2000; Goldammer et al., 2004; Azawi et al., 2008; Davies et al., 2008). With the agreement of these data, we detected overexpression of all five examined TLR genes, particularly TLR2 (26.4 fold) and TLR4 (12.4 fold), in infected uterine tissues with endometritis compared to the uninfected ones.
Conclusion
Our data demonstrated that the variations in nucleotides and amino acids sequence between Bubalus bubalis with and without endometritis are not associated with endometritis. In addition, there is an overexpression of examined TLRs genes especially TLR2 and TLR4 in animals with endometritis compared to animals without endometritis specifically in uterine tissue.
Acknowledgements
The authors would like to thank prof. Ostrowski (Department of Oncological Genetics, Maria Sklodowska-Curie Memorial Cancer Center and Institute of Oncology, Warsaw, Poland) for providing all facilities to carry out his work.
Conflict of Interest
The authors declared no conflict of interest.
References
Transcriptional profile of endometrial TLR4 and 5 genes during the estrous cycle and uterine infection in the buffalo (Bubalus bubalis). Vet. Res. Commun. 38(2): 171-176. http://dx.doi.org/10.1007/s11259-014-9594-1




