Journal of Infection and Molecular Biology
Research Article
Bacteremia Prediction by Inflammatory Factors and Recent Trend in Drug Resistance of Bacteria Isolated from Blood Stream Infection
Muhammad Sohail1, 2*, Qamar Sultana2, 6, Karam Rasool2, 4, Samreen Sarwar3, Abdul Basit2,5, Muhammad Khalid2
1Department of Microbiology and Molecular Genetics, University of the Punjab, Lahore, 54000, Pakistan; 2Chughtais Lahore Lab, Lahore, 54660, Pakistan; 3University of Health Sciences Lahore, 54000, Pakistan; 4Department of Microbiology, University of Veterinary and Animal Sciences, Lahore, Pakistan; 5Forensic Sciences, University of Veterinary and Animal Sciences, Lahore, Pakistan; 6Department of Pathology, Sir Ganga Ram Hospital, Queen’s Road, Lahore, Pakistan.
Abstract | Bacteremia is associated with high rates of morbidity and mortality. For early diagnosis of bacteremia; prior use of antibiotics, suboptimal volume of blood sample and improper microbiological techniques are strongly condemned. In time-diagnosis of bacteremia can reduce the antibiotic resistance, hospital stay and cost to the treatment. Blood samples from patients with history of fever in conjunction with clinical signs and symptoms of bacteremia (from July 2013 to July 2014) were processed using BACTEC 9050 automated system. ESR (erythrocyte sedimentation rate), CRP (C- reactive protein) and TLC (total leukocyte count) were also measured, respectively. Out of 3559 samples, 525 (14.75%) samples were positive for bacterial growth. Among positive blood cultures, the pathogens isolated were coagulase negative Staphylococcus aureus (36.38 %), E. coli (18.28%), Methicillin sensitive Staphylococcus aureus (MSSA) (7.04%), Strept. fecalis (5.9%), Salmonella typhi (6.09%), Candida species (4.95%), Pseudomonas species (4.38%), Klebsiella pneumonia (4%), Acinetobacter species (3.42%), Salmonella paratyphi A (2.85%), Methicillin resistant Staphylococcus aureus (MRSA) (2.67%), Citrobacter species (2.47%), Streptococcus species other than Strept. faecalis (1.52%), Xanthomonas maltophilia (0.57%), Vibrio species (0.19%), Yersinia species (0.19%) and Klebsiella oxytoca (0.19%). ESR, CRP and TLC showed poor correlation with bacteremic patients. In addition, increased antimicrobial resistance was observed in all pathogens isolated. ESR, CRP and TLC were considered as preliminary indicator of infections having low specificity and sensitivity in blood stream infections.
Keywords | Antimicrobial susceptibility testing, Bacteremia, Total leukocyte count, Erythrocyte sedimentation rate, C-reactive protein
Editor | Tahir Yaqub, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | July 02, 2015; Revised | October 11, 2015; Accepted | October 14, 2015; Published | November 14, 2015
*Correspondence | Muhammad Sohail, 10-Jail Road, Chughtais Lahore Lab, Lahore, Pakistan; Email: drsohailmmg@gmail.com
Citation | Sohail M, Sultana Q, Rasool K, Sarwar S, Basit A, Khalid M (2015). Bacteremia prediction by inflammatory factors and recent trend in drug resistance of bacteria isolated from blood stream infection. J. Inf. Mol. Biol. 3(3): 75-80.
DOI | http://dx.doi.org/10.14737/journal.jimb/2015/3.3.75.80
ISSN (Online) | 2307-5465; ISSN (Print) | 2307-5716
Copyright © 2015 Sohail et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
International sepsis guidelines recommend blood culture and sensitivity testing before initiation of antimicrobial chemotherapy for targeted treatment (Dellinger et al., 2013). Blood culture plays key role in antibiotic stewardship program (Standiford et al., 2012). Recovery of pathogens from blood is considerably low in resource limited settings i.e., 30 – 40% in severe sepsis due to lack of adherence to standard operating procedures (SOPs) of microbiology laboratory and inappropriate sample collection practices (Schmitz et al., 2013). The Clinical and Laboratory Standard Institute (CLSI) recommends sample volume of 60 mL for recovery of pathogens from blood culture (Weinstein et al., 1997) but in clinical lab settings of Pakistan, 20 – 30 mL blood is hardly sent to laboratory for diagnosis of bacteremia. Another important factor that leads to poor recovery of pathogens from blood is empirical antibiotic therapy. The recovery can be optimized by using antibiotic neutralizing substances in blood culture bottles if empirical therapy is crucial.
Some inflammatory markers are linked to generalized bacterial infections including bacteremia (Feldman et al., 2013). The CRP (C-Reactive protein) and ESR (Erythrocyte Sedimentation Rate) are inflammatory markers which may predict some of the pathological conditions (Keenan et al., 2008). The increase in total leucocyte count (TLC) in fever is also an important indicator of bacterial infections including bacteremia (Jaffe and Fleisher, 1991).
This prospective study was conducted to determine relationship between inflammatory markers (CRP and ESR), TLC and bacteremia. Prevalence of different pathogens in bacteremia and recent trend in the antimicrobial activity of reported pathogens has also been evaluated in this study.
Materials and Methods
A total of 3559 blood culture samples were received in microbiology department of Chughtais Lahore lab with clinical sign and symptoms of bacteremia. Blood culture samples were received in the BACTEC blood culture bottles (Becton, Dickinson and Company, Sparks, MD, USA) from more than 100 sample collection centers in Pakistan. Proper transport of samples was ensured with careful monitoring of transport conditions. Each blood culture bottle was incubated for five days before being discarded as negative. All positive blood culture bottles were further sub-cultured for identification of pathogen and antimicrobial susceptibility testing. Positive blood cultures were inoculated on Blood and MacConkey agar (Oxoid, UK) and incubated at 37°C for 24 hours. The organisms were identified using standard microbiological procedures. Bacteremia with single pathogen was included in this study while others co-infections were not included in present study. CLSI 2013 guidelines were followed for antimicrobial susceptibility testing (AST). AST was performed on Mueller-Hinton agar (Oxoid, UK) using Kirby Bauer disk diffusion technique. Commercially available drugs with defined concentrations were used for isolates as per recommended standard (CLSI 2013). Antimicrobial drugs used for gram positive and gram negative bacterial isolates were ampicillin (AMP) (30μg), amoxicillin (AML) (25μg), amoxicillin/clavulanic acid (AMC) (30μg), ampicillin/sulbactum (SAM) (30μg), cefipime (FEP) (30μg), cefotaxime (CTX) (30μg), cefoxitin (FOX) (30μg), cefuroxime (CXM) (30μg), ceftazidime (CAZ) (30μg), ceftriaxone (CRO) (30μg), tigecycline (TGC) (15μg), cephalexin (CL) (30μg), cephradine (CE) (30μg), cefaclor (CEC) (30μg), cefixime (CFM) (30μg), imipenem (IMI) (10μg), meropenem (MEM) (10μg), vancomycin (VA) (30μg), amikacin (AK) (30μg), gentamycin (CN) (30μg), tobramycin (TOB) (30μg), azithromycin (AZM) (30μg), erythromycin (E) (30μg), doxycycline (DO) (30μg), ciprofloxacin (CIP) (10μg), levofloxacin (LEV) (10μg), clindamycin (DA) (10μg), fusidic acid (FD) (50μg), linezolid (LZD) (30μg), cefoperazone (CFP) (30μg), aztreonam (ATM) (30μg), nalidixic acid (NA) (30μg), ofloxacin (OFX) (5μg), moxifloxacin (MXF) (5μg), trimethoprim/sulfamethoxazole (SXT) (25μg), cefoperazone/sulbactam (SCF) (30μg) and piperacillin/tazobactam (TZP) (110μg). The minimum inhibitory concentration (MIC) of colistin (0.064-1024 mg/L), polymyxin B (0.064-1024 mg/;) for Acinetobacter species and Vancomycin (0.016-256 mg/L) for Staphylococci and Streptococci species was studied by E-test method (Biomerieux, France). The MIC was interpreted to be a value where growth inhibition zone meets the E-test strip excluding small colonies in the ellipse (Falagas et al., 2008). E. coli (ATCC® 25922), Pseudomonas aeruginosa (ATCC® 27853), E. faecalis (ATCC®229212) and Staphylococcus aureus (ATCC® 29213) were used as reference strains according to CLSI 2013 standard for susceptibility testing.
Inflammatory markers (ESR and CRP) were performed for a defined number of negative and positive blood culture samples as per patient request form. The selection of samples was random. CRP was measured quantitatively in serum using Immunoturbidimetric technology on the ARCHITECT c Systems (Architect c8000, Abbott Laboratories). TLC was performed using CELL-DYN 1800 Hematology Analyser and ESR was performed by Westergren method.
Results
Samples were collected from July 2013 to July 2014 from different cities of Pakistan. In this study, 14.75% (N= 525) blood cultures were positive for single pathogens. The pathogens isolated from positive blood cultures were Coagulase negative Staphylococcus aureus (CoNS) (36.38%), E. coli (18.28%), Methicillin sensitive Staphylococcus aureus (MSSA) (7.04%), Salmonella typhi (6.09%), Streptococcus faecalis (5.9%), Klebsiella pneumoniae (4%), different Candida species (4.95%), Pseudomonas spp. (4.38%), Acinetobacter spp. (3.42%), Salmonella paratyphi A (2.85%), Methicillin resistant Staphylococcus aureus (MRSA) (2.67%), Citrobacter spp. (2.47%), Streptococcus species other than Strept. pyogenes and Strept. faecalis (0.95%), Xanthomonas maltophilia (0.57%) and Strept. pyogenes (Table 1). These results indicated that Coagulase negative Staphylococcus (CoNS) is the most commonly isolated pathogen from bacteremic patients followed by E. coli, MSSA, S. typhi, Strept. faecalis and Klebsiella pneumoniae.
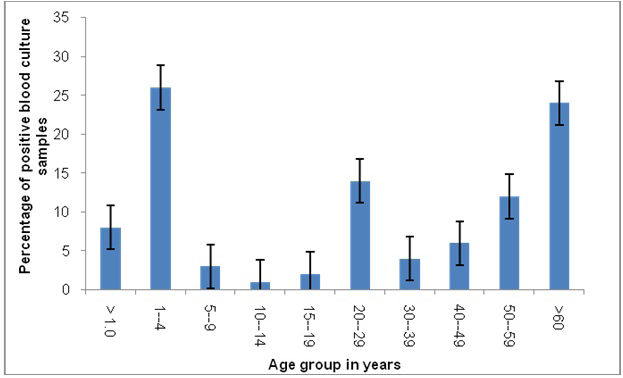
Prevalence of positive blood cultures was 38% and 62% in female and male patients, respectively. Age wise prevalence showed that age group less than 5 years and more than 60 years are most susceptible to bacteremia. Age groups
Table 1: Prevalence of bacteremic pathogens
|
Gram negative bacteria |
Gram positive bacteria |
Yeast |
|||
|
E. coli |
99(18.28%) |
CoNS |
191(36.38%) |
Candida |
26(4.95%) |
|
K. pneumoniae |
21(4%) |
MSSA |
37(7.04%) |
||
|
S. typhi |
32(6.09%) |
St. fecalis |
30(5.71%) |
||
|
Acinetobacter |
18(3.42%) |
MRSA |
14(2.67%) |
||
|
S. paratyphi A |
15(2.85%) |
Streptococcus |
5(0.97%) |
||
|
P. aeruginosa |
14(2.67%) |
St. pyogenes |
3(0.95%) |
||
|
Citrobacter |
3(2.47%) |
||||
|
Xanthomonas maltophilia |
1(0.19%) |
||||
|
Vibrio |
2(0.38%) |
||||
|
Yersinia |
1(0.19%) |
||||
|
pseudomonas |
9(1.71%) |
||||
<1, 1-4, 5-9, 10-14, 15-19, 20-29, 30-39, 40-49, 50-59 and >60 years have positive blood culture rate of 8%, 26%, 3%, 1%, 2%, 14%, 4%, 6%, 12% and 24% respectively. Middle age group (20-29 years) is also among most affected age groups (Figure 1).
Among negative blood cultures, ESR, CRP and TLC were performed on 155, 369 and 743 samples respectively, while for positive blood cultures these tests were performed on 31, 98 and 227 samples, respectively. In is clear from results that ESR was almost equally high in positive and negative blood culture samples (Figure 2). CRP levels were high for negative than positive blood culture samples indicating non-bacteremic specific CRP elevations. TLC values correlated with blood culture results. TLC is higher for positive blood culture and normal for negative blood culture.
The percentage of resistance to various antimicrobials was determined against isolated pathogens. Isolates of E. coli showed 100% resistance to ampicillin and amoxicillin while 96% resistance to augmentin and sulbactum. Resistance to imipenem, amikacin, sulzone and tazocin was 30%, 17%, 19% and 22% respectively. These relative low resistance percentages make these drugs suitable for treatment of multidrug resistant and ESBL producing E. coli. Citrobacter, Enterobacter and Vibrio species were sensitive to most of the drugs tested.
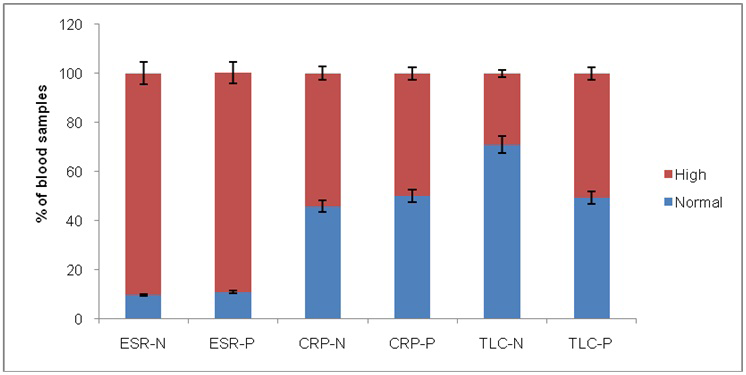
Figure 2: Bar chart showing the percentage of normal and high ESR, CRP and TLC values in positive and negative blood samples
ESR-N: ESR in negative blood culture; ESR-P: ESR in positive blood culture; CRP-N: CRP in negative blood culture; CRP-P: CRP in positive blood culture; TLC-N: TLC in negative blood culture; TLC-P: TLC in positive blood culture
There was only single isolate of Klebsiella oxytoca and therefore does not truly depict antibiogram for all K. oxytoca infections. In hospital acquired infections, Acinetobacter species were found to be resistant to most of available drugs. Percentage resistance of Acinetobacter species to tigecycline, doxycycline and sulzone was 17%, 72% and 11%, respectively (Table 2). In these cases, minimum inhibitory concentration (MIC) of colistin and polymyxin B was determined. MIC of colistin and polymyxin B was performed by E-test method. All isolates of Acinetobacter species were sensitive to colistin and polymyxin B.
Two isolates of CoNS were resistant to Clindamycin while most of the isolates were sensitive to penicillins, cephems, aminoglycolides and macrolides (Table 3). MRSA was found to be resistant to most of available drugs and results supported the use of linezolid and vancomycin for treatment of MRSA infection. For Pseudomonas species, tazocin
Table 2: Percentage resistance of gram negative bacteremic pathogens to antimicrobial agents (NT, not tested
|
Pathogens isolated / Antimicrobial drug used |
AMC |
SAM |
FEP |
CFP |
CXM |
CRO |
CFM |
IMI |
ATM |
AK |
DO |
CIP |
LEV |
MOX |
SXT |
TZP |
|
E. coli |
96 |
96 |
77 |
85 |
88 |
85 |
84 |
17 |
80 |
30 |
79 |
73 |
73 |
70 |
83 |
22 |
|
Klebsiella pneumoniae |
100 |
100 |
90 |
95 |
100 |
95 |
95 |
19 |
95 |
67 |
71 |
57 |
57 |
57 |
90 |
33 |
|
S. typhi |
19 |
19 |
9 |
9 |
NT |
9 |
9 |
3 |
NT |
3 |
6 |
56 |
56 |
56 |
59 |
NT |
|
Acinetobacter |
100 |
100 |
100 |
100 |
89 |
100 |
100 |
94 |
89 |
83 |
72 |
88 |
88 |
88 |
83 |
83 |
|
S. paratyphi A |
7 |
7 |
NT |
NT |
NT |
NT |
NT |
NT |
NT |
NT |
27 |
54 |
54 |
54 |
47 |
NT |
|
Citrobacter |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
33 |
33 |
0 |
0 |
0 |
|
Enterobacter |
100 |
100 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Xanthomonasmaltophilia |
NT |
NT |
N T |
NT |
NT |
NT |
NT |
NT |
NT |
NT |
33 |
NT |
33 |
NT |
33 |
NT |
|
Vibrio species |
100 |
100 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Yersinia Species |
100 |
100 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
100 |
0 |
|
Klebsiella oxytoca |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
0 |
100 |
100 |
100 |
100 |
100 |
100 |
0 |
100 |
|
Enterococcus spp. |
100 |
100 |
100 |
100 |
100 |
100 |
100 |
0 |
100 |
100 |
100 |
100 |
100 |
100 |
0 |
0 |
Table 3: Percentage resistance of gram positive bacteremic pathogens to antimicrobial agents
|
Pathogen isolated / Antimicrobial drug used |
AMC |
CFP |
CTX |
FOX |
CRO |
IMP |
ATM |
CN |
TOB |
DO |
CIP |
LEV |
OFX |
TZP |
LZD |
DA |
FD |
|
CoNS |
40 |
40 |
40 |
39 |
40 |
40 |
4 |
40 |
46 |
41 |
50 |
50 |
16 |
NT |
2 |
31 |
40 |
|
MSSA |
0 |
0 |
0 |
0 |
0 |
0 |
NT |
11 |
16 |
16 |
24 |
24 |
5 |
NT |
0 |
16 |
14 |
|
St .fecalis |
37 |
NT |
NT |
NT |
NT |
NT |
NT |
80 |
13 |
73 |
73 |
73 |
33 |
NT |
0 |
13 |
10 |
|
P. aeruginosa |
NT |
NT |
NT |
NT |
NT |
7 |
14 |
28 |
28 |
36 |
0 |
0 |
0 |
7 |
NT |
NT |
NT |
|
MRSA |
93 |
93 |
93 |
93 |
93 |
86 |
14 |
71 |
86 |
43 |
86 |
86 |
28 |
NT |
0 |
43 |
43 |
|
St. pyogenes |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
100 |
50 |
0 |
50 |
50 |
0 |
NT |
0 |
50 |
50 |
|
Pseudomonas sp. Other than P. aeruginosa |
0 |
0 |
0 |
0 |
0 |
11 |
45 |
11 |
11 |
11 |
11 |
11 |
11 |
11 |
NT |
NT |
NT |
NT: not tested; MRSA: Methicillin resistant Staphylococcus aureus; MSSA: Methicillin sensitive Staphylococcus aureus; CoNS: Coagulase negative Staphylococcus
gave best results. MIC of vancomycin was determined by E-test and results were interpreted as per CLSI 2013 guidelines. All of the MSSA, MRSA, Strept. fecalis, Strept. pyogenes, CoNS and Strept. pneumoniae showed sensitivity to vancomycin.
Discussion
Sepsis is detected by blood culture which is considered as gold standard for sepsis throughout world (Martin et al., 2003). Improper sampling technique, timing of sample collection from patient, suboptimal volume of blood, time of incubation, sample transportation conditions, use of antibiotics prior to culture blood sample and improper microbiological techniques are major challenges in timely and accurate diagnosis of pathogens responsible for sepsis. In our study, an average of 10 mL sample volume was used. 14.75% positive and 85.25% negative blood culture rates correlated with a study conducted by Gohel et al. (2014). The improper sample transport conditions observed in distant sample collection centers, can have impact by increasing false negative results. The incidence of gram positive and gram negative pathogens in blood culture was 53 % and 47 % respectively. These results were similar to the results reported by Ayobola et al. (2011). Our study showed that gram positive pathogens are more prevalent bacteremia pathogens than gram negative organisms in this setting. The high rate of gram positive infections can be due to use of prosthetic devices and pathogenesis of gram positive pathogens involving deep tissue infections.
Staphylococci were isolated in 47% of the cases, out of which 36% were CoNS in present study. The isolation of CoNS in our study is in accordance with various other studies such as Arora and Devi (2007), Desai and Malek (2010) and Jones and his coworkers (Karlowsky et al., 2004). E. coli was the predominant gram negative isolate (18%) in our study. Similar rate was reported in a study conducted by Falagas et al. (2008). Similar frequencies of Pseudomonas, Salmonella, Acinetobacter, Candida, Moraxella and Streptococcus species were obtained as reported by Gohel and his colleagues (Gohel et al., 2014).
Resistance to oxacillin (83%) was found to be greater than previously reported studies and showed alarming emergence of increasingly resistant superbugs in bloodstream infections. The antibiotics which were least resistant, and can be drug of choice are vancomycin, macrolids, clindamycin and aminoglycosides for Staphylococcus species while doxycyclin, fluorequinalines, colistin and cephems for enterobacteriaceae.
Continuous assessment of CRP, TLC and ESR alone or along with other parameters give indication of multiple infections including bacteremia (Altrichter et al., 2010). Our study showed that these parameters give non-parallel indication of bacteremia but continuous assessment of these parameters can be helpful for prediction of bacteremia when there is asymptomatic fever history. CRP, ESR and TLC are disturbed in the multiple condition including urinary tract infections, gastrointestinal infection and blood stream infections (Cengić et al., 2001). In this study, ESR was elevated in 90% of positive cases and negative cases and therefore showed that ESR is highly non-specific for bloodstream infections. CRP was also elevated in 50% of positive cases while TLC was elevated in 29% negative cases and 50% positive cases. The data analysis showed that these parameters are useful along with fever and other clinical indications but are not as reliable and specific as considered.
Conclusion
Among gram positive pathogens, CoNS was prominent pathogens followed by MSSA, Strept. fecalis and MRSA. Escherichia. coli is predominant pathogen followed by Klebsiella, Salmonella and Acinetobacter. Overall resistance of bacteremic pathogens to antimicrobial drugs was found to be elevated when compared to past studies. ESR, CRP and TLC are not strong indicators of blood stream infections alone but these can be useful indicators of bacteremia along with continuous fever and other clinical indications.
Conflicts of interest
There is no conflict of interest regarding this work.
Acknowledgements
This research is non-funded and part of routine work conducted at Chughtais Lahore Lab and is gratefully acknowledged.
Authors’ contribution
Qamar Sultana and Muhammad Sohail came up with concept, designed the Study and gave administrative, technical, and material support; Muhammad Sohail; Abdul Basit and Muhammad Khalid collected the data; Samreen sarwar and Karam Rasool analysised and interpred data while Karam Rasool, Samreen Sarwar, and Abdu Basit drafted the manuscript. Qamar Sultana and Abdul Basit critically revised the manuscript for important intellectual content and Karam Rasool performed statistical analysis. The whole procedure of the study was supervised by Qamar Sultana and Samreen Sarwar.
References