Journal of Infection and Molecular Biology
Research Article
Prevalence of Aflatoxigenic Aspergillus in Food and Feed Samples from Karachi, Pakistan
Fariha Ibrahim1a, Hina Jalal1a, Abdul Basit Khan1, 2*, Muhammad Asif Asghar2, Javed Iqbal2, Aftab Ahmed2, Ghufrana Nadeem1
1Deparment of Microbiology, Jinnah University for Women, 5-C Nazimabad, Karachi, Pakistan; 2Food and Feed Safety Laboratory, Food and Marine Resources Research Centre, PCSIR Laboratories Complex, Shahrah-e-Salimuzzaman Siddiqui, Off University Road, Karachi−75280, Pakistan.
Abstract | Aflatoxins are naturally occurring toxic metabolites synthesized by certain species of Aspergillus, in particular Aspergillus flavus and Aspergillus parasiticus. Several types of aflatoxins such as B1, B2, G1, G2, M1, and M2 are produced by these fungal strains. Among these, aflatoxin B1 is the most toxic to mammals and induces cell injury. Epidemiological, clinical and experimental studies revealed that aflatoxins are toxic and carcinogenic to mammals. This study was designed to observe the total fungal count and the prevalence of aflatoxin-producing Aspergillus using polymerase chain reaction (PCR) in local food and feed products. A total of 102 food and feed product samples were randomly collected and primarily analyzed to observe the total yeast and mould count. Subsequently, Aspergillus species were isolated and screened for ver-1, apa-2, omt-1 genes of aflatoxin biosynthetic pathway using PCR assay. Thin layer chromatography (TLC) was performed to confirm the synthesis of aflatoxin in PCR-positive strains. It was observed that all tested samples were found contaminated with yeast and moulds having highest count in cotton seed cake samples. A total of 102 Aspergillus strains were isolated, in which 09 were positive to all three targeted genes, wherein 06 were A. flavus and 03 were A. parasiticus positive. All PCR positive strains produced aflatoxin B1 and B2, but not G1 and G2 when analyzed by TLC. It was observed that Aspergillus species were one of the most common contaminants in food and feed samples. Aflatoxigenic strains of Aspergillus were also frequent and it is important to devise strategies at local levels to control these aflatoxin producing strains at pre- and post-harvest stages.
Keywords | Aspergillus, Aflatoxins, omt-1, ver-1, apa-2
Editor | Tahir Yaqub, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | March 30, 2016; Accepted | May 12, 2016; Published | May 26, 2016
*Correspondence | Dr. Abdul Basit Khan, Department of Microbiology, Jinnah University for Women, 5-C, Nazimabad, Karachi, Pakistan; E-mail: basitanalyst@gmail.com
Citation | Ibrahim F, Jalal H, Khan AB, Asghar MA, Iqbal J, Ahmed A, Nadeem G (2016). Prevalence of aflatoxigenic Aspergillus in food and feed samples from Karachi, Pakistan. J. Inf. Mol. Biol. 4(1): 1-8.
a These authors contributed equally to this work.
DOI | http://dx.doi.org/10.14737/journal.jimb/2016/4.1.1.8
ISSN (Online) | 2307-5465; ISSN (Print) | 2307-5716
Copyright © 2016 Ibrahim et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Fungal contamination is one of the major contributors of food products spoilage and grains deterioration during storage, rendering them unfit for human consumption. Some of these contaminants, for instance, few species of Aspergillus and Fusarium produce mycotoxins in many food products. The quality of food and related products, such as seeds/grains, vegetables, fruits and processed products, are significantly deteriorated by these toxigenic strains (Lee et al., 2007).
Different types of fungal toxins have been discovered so far, presenting different structural diversity, chemical composition and physicochemical properties. Among all of these, aflatoxins (AFs) are the most commonly occurring and extensively studied mycotoxin in food and feed commodities. They are produced as secondary metabolites and 4 types i.e., aflatoxin B1 (AFB1), B2 (AFB2), G1 (AFG1) and G2 (AFG2) are the most important with reference to mutagenic, teratogenic, and immunosuppressive capabilities (Lereau et al., 2012). They are classified in group I as first class carcinogens, mutagens and immunosuppressive agents by International Agency for Research on Cancer (IARC Monograph, 2002).
Several studies revealed that A. flavus and A. parasiticus are primarily involved in the production of AFs (Ghadeer and Al-Delamiy, 2012). These two fungal species, especially A. flavus, have been reported in most of the cases of aflatoxin contamination (Varga et al., 2011). The members of Aspergillus species are capable to establish themselves in a variety of food products and in diverse environmental conditions. However, typical hot and humid atmosphere and substandard storage conditions are required to synthesize aflatoxins in agricultural products (Atanda, 2013).
There are several conventional and chromatography-based techniques, for example thin layer chromatography (TLC), gas chromatography (GC), high performance liquid chromatography (HPLC), etc., are available for the detection of aflatoxins and aflatoxigenic moulds (Bacaloni et al., 2008). However, Polymerase Chain Reaction (PCR) has an edge on other methods developed to screen aflatoxigenic fungi. A number of genes such as afl R, nor-1, omt-1, ver-1 involved in the biosynthetic pathway of aflatoxin synthesis, have been targeted in this regard (Criseo et al., 2001). Aflatoxins (AFs) contamination in food and feed products is a major public health problem in developing countries (Eaton and Gallagher, 1994). It has been estimated that more than 5 billion people are at risk of continuous exposure to aflatoxins through consumption of contaminated food products in developing countries (Strosnider et al., 2006). So, the objective of the present study was to observe the fungal load of different food and feed products on one hand and to examine the prevalence of aflatoxigenic Aspergillus strains by using polymerase chain reaction on the other hand.
Materials and Methods
Chemicals and Reagents
Aflatoxin B1, B2, G1 and G2 standards were obtained from Biopure (Vienna, Austria). Potato dextrose agar (PDA) and Czepak Dox liquid medium (CZK) were purchased from Oxoid limited (Basingstoke, Hampshire, England). Acetone, acetonitrile (ACN), benzene (C6H6), chloroform (CHCl3), ethanol (EtOH), 2-propanol, sodium chloride (NaCl), 2-mercaptoethanol, peptone and agarose were obtained from Merck (Darmstadt, Germany). Hydrochloric acid (HCl), potassium acetate (K(CH3COO)2), potassium sulphate (K2SO4), sodium hydroxide (NaOH), sulphuric acid (H2SO4), sodium sulphate (Na2SO4), sodium dodecyl sulphate (SDS), Tris base, EDTA and ethidium bromide were procured from Sigma-Aldrich (St. Louis-MO, USA). GoTaq® Green master mix for PCR was acquired from Promega (Medison-WI, USA). DNA primers were synthesized by Integrated DNA technologies (IDT) (Coralville-Iowa, USA).
Sample Collection
A total of 102 samples of food, feed and agricultural commodities comprising rice (n=18), maize (n=23), wheat (n=05), red chillies (n=06), composite spices (n=08), poultry feed (n=28) and cotton-seed cake (n=14) were collected from local markets of Karachi-Pakistan during the year 2013. Collected samples were separately minced using a sample grinder and a homogeneous sample was prepared. Samples were stored at -20oC in air-tight plastic bags till further processing.
Total Yeast and Mould Count
A method described in bacteriological analytical manual was employed to enumerate total yeast and mould count (Valerie et al., 2001). Briefly, 25g of each homogenized sample was weighed and initially diluted in 225ml of 0.1% peptone water to achieve 1:10 dilution followed by serial dilution to 1:103 or 1:104 in 0.1% peptone water. One milliliter of each dilution was inoculated to Potato Dextrose Agar incubated at 25°C for 5 days. After incubation, representative fungal colonies were counted and colony forming unit per gram (CFU/g) was calculated. Colonies of suspected Aspergillus were subcultured and examined for species identification. The isolation and characterization of Aspergillus species was based on colony color, growth pattern and microscopic characteristics.
Polymerase Chain Reaction to Detect Aflatoxigenic Aspergillus Strains
DNA extraction was performed according to the method described by Petra and Vytrasova (2011) with slight modifications. Briefly, Aspergillus strains were inoculated in 100ml Czapek Dox liquid medium and incubated at 28°C for 5-7 days. After incubation, content of flask was filtered through Whatman filter paper 4 and fungal mycelia were dried in oven at 50°C for 24 hours. Total DNA was extracted by incubating 70mg of fungal mycelium with 700µL DNA extraction buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, 100 mM NaCl, 10 mM 2-mercaptoethanol and 1% SDS) at 65°C for 10 minutes. Thereafter, 200µL of 5M potassium acetate was added and DNA suspension was incubated again at -20 ͦ C for 10 minutes followed by centrifugation at 12000rpm for 10 minutes. Subsequently, 400ul of supernatant was diluted with equal volume of 2-propanol and centrifuged at 6000rpm for 10 minutes. The supernatant was removed and pellet was washed with 70% (v/v) ethanol in two steps. Finally, DNA containing pellet was dried and resuspended in 50µL of Tris-EDTA (TE) buffer.
Polymerase chain reaction (PCR) was used to screen Aspergillus species for the presence of 3 different genes responsible for the synthesis of enzymes involved in Aflatoxin biosynthetic pathway. Primers sequences and PCR conditions
Table 1: List of target genes and their respective primer sequences
|
S.No. |
Target Gene |
Primer name |
Sequences (5’-3’) |
Length of PCR Product (bp) |
|
1 |
ver-1 |
VER-496 |
ATGTCGGATAATCACCGTTTAGATGGC |
895 |
|
VER-1391 |
CGAAAAGCGCCACCATCCACCCCAATG |
|||
|
2 |
apa-2 |
APA-450 |
TATCTCCCCCCGGGCATCTCCCGG |
1032 |
|
APA-1482 |
CCGTCAGACAGCCACTGGACACGG |
|||
|
3 |
omt-1 |
OMT-208 |
GGCCCGGTTCCTTGGCTCCTAAGC |
1024 |
|
OMT-1232 |
CGCCCCAGTGAGACCCTTCCTCG |
were employed as described earlier by Shapira et al. (1996). The sequences of primers are listed in Table 1.
PCR assay was performed in 25µL of a reaction mixture that contained 2.5µL of extracted genomic DNA, 12.5µL of master mix, 0.5µL of each forward and reverse primer. The final volume was made up to 25µL with nuclease free water. PCR was started by initial denaturation at 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 60 seconds, annealing at 65°C for 120 seconds and extension at 72°C for 120 seconds. The process was concluded at 72°C for 5 minutes as final extension. PCR products were resolved on 1% agarose gel and stained with ethidium bromide. After electrophoresis, gel was removed from the gel-casting platform, exposed to UV transillumination and photographs were taken.
Aflatoxin Detection by Thin Layer Chromatography (TLC)
Spore suspension was prepared according to the method described by Ghadeer and Al-Delamiy (2012). Briefly, the growth of each suspected Aspergillus species in petri plates was scraped with 10ml sterile distilled water and suspended into 90 ml of sterile distilled water. The spore suspension was further diluted up to 107 spores/ml with sterile distilled water.
The sample for thin layer chromatography was prepared as reported earlier by Criseo et al. (2001) with slight modifications. Briefly, 0.1 ml of spore suspension was inoculated in 100ml of czapek dox medium and incubated at 25°C ± 2oC for 10 days. Thereafter, the whole suspension was blended at 5000 rpm for 2 min. The mycelia were separated by filtration and re-blended with 50 ml of chloroform. The mycelium blend was again filtered; extracts were combined and dried in a Rota vapor under N2 atmosphere. Aflatoxins from filtrate were extracted with 100 ml of chloroform. Finally, chloroform layer was separated over anhydrous sodium sulphate (ca. 1g). The dried sample extracts were run on TLC for aflatoxins detection and quantification.
Thin layer chromatography (TLC) was performed as described by Asghar et al. (2014)ried extracts were suspended in 100ml of benzene-acetonitrile (98:2; v/v) and shaken well. Samples were spotted as 2, 5 and 10µL. The AF standards were separately loaded on the TLC plates on imagining line about 4 cm from bottom edge. TLC plates were developed in chloroform: xylene: acetone (6:3:1; v/v) and was observed under long wavelength UV light (λ= 254nm and 366nm) in UV visualizer. The retention factor (Rf) of each AFs (B1, B2,G1 or G2) was calculated according to the following equation.
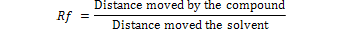
All positive results of naturally existing AFs in the tested samples were confirmed by spraying the TLC plates using 50% (v/v) H2SO4.
Statistical Analysis
The Student’s t test (p < 0.05 as the minimal level of significance unless indicated otherwise) was used to analyze statistical data and all values were expressed as the means ± SD.
Results
Total Yeast and Mould Count
A total of 102 food and feed samples were obtained from Karachi-Pakistan during the year of 2013. All samples were initially tested to determine the total load of yeast and moulds and cfu/g was calculated. Average cfu/g of each commodity was calculated and represented as bar graph as shown in Figure 1.
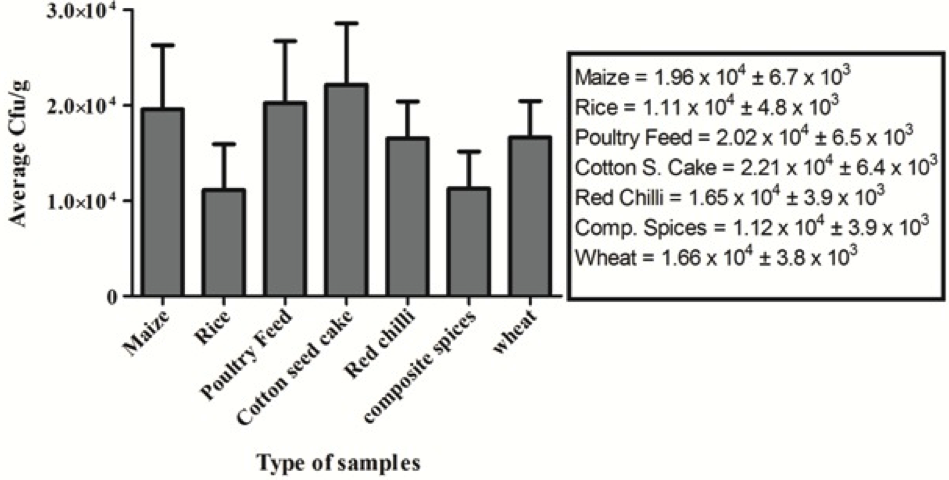
Figure 1: >Average total yeast and mould count of different food commodities; Triplicate analyses were made for all measurements. Indicated values are means (n = 3) ± SD
(A & B) omt-1gene (Amplified 1,024bp); (C & D) apa-2 gene (Amplified 1,032bp); (E) ver-1 gene (Amplified 895bp) FH07: A. flavus; FH25: A. flavu; FH39: A. flavus; FH44: A. parasiticus; FH74: A. flavus; FH72: A. flavu; FH65: A. parasiticus; FH56: A.parasiticus; FH55: A. flavus; SM: 1kb DNA size marker; -ve control containing master mix without samples
Isolation of Aspergillus Species
In this study, 102 Aspergillus species were isolated from various food and feed samples consisting of composite spices, cotton-seed cake, maize, poultry feed, red chillies, rice and wheat. These species were identified according to growth, colony color and microscopic characteristics. In the present study three most common Aspergillus species were found including, Aspergillus flavus (n=50), Aspergillus parasiticus (n=06) and Aspergillus niger (n=46), respectively. Aspergillus flavus exhibited olive to lime green colonies with cream reverse, rough conidia and septate hyphae. Whereas, Aspergillus parasiticus isolates were characterized by the presence of dark green colonies with hyaline hyphae. However, Aspergillus niger showed black colonies, with pale yellow reverse, rough conidia and hyaline hyphae.
Polymerase Chain Reaction to Detect Aflatoxigenic Aspergillus Strain
DNA was amplified through PCR according to the described conditions. The PCR was conducted using the sets of three primers specifically for ver-1, apa-2, and omt-1 genes targeted the Aspergillus species DNA. The expected
Table 2: Results of PCR for the determination of target genes
|
S. No. |
Food Commodities |
Strains Designation |
Aspergillus Species |
PCR target genes |
||
|
ver-1 |
apa-2 |
omt-1 |
||||
|
1 |
Cotton Seed Cake |
FH07 |
A.flavus |
+ |
+ |
+ |
|
2 |
Maize |
FH25 |
A.flavus |
+ |
+ |
+ |
|
3 |
Maize |
FH44 |
A.parasiticus |
+ |
+ |
+ |
|
4 |
Cotton Seed Cake |
FH39 |
A.flavus |
+ |
+ |
+ |
|
5 |
Poultry Feed |
FH55 |
A.parasiticus |
+ |
+ |
+ |
|
6 |
Poultry Feed |
FH56 |
A.parasiticus |
+ |
+ |
+ |
|
7 |
Maize |
FH65 |
A.flavus |
+ |
+ |
+ |
|
8 |
Poultry Feed |
FH72 |
A.flavus |
+ |
+ |
+ |
|
9 |
Poultry Feed |
FH74 |
A.flavus |
+ |
+ |
+ |
Table 3: Aflatoxins production in Aspergillus species. Triplicate analyses were made for all measurements. Indicated values are means (n = 3) ± SD
|
S. No. |
Food Commodities |
Strains Designation |
Aspergillus Species |
Conc. Of Aflatoxins (µg/kg) |
|
|
AFB1 |
AFB2 |
||||
|
1 |
Cotton Seed Cake |
FH07 |
Flavus |
200.27 ± 5.54 |
20.07 ± 0.97 |
|
2 |
Maize |
FH25 |
Flavus |
460.32 ±7.23 |
56.02 ± 1.32 |
|
3 |
Maize |
FH44 |
parasiticus |
500.82 ± 6.46 |
52.65 ± 1.56 |
|
4 |
Cotton Seed Cake |
FH39 |
flavus |
320.98 ± 3.21 |
37.63 ± 0.88 |
|
5 |
Poultry Feed |
FH55 |
flavus |
275.22 ± 4.71 |
27.51 ± 1.17 |
|
6 |
Poultry Feed |
FH56 |
parasiticus |
250.38 ± 3.38 |
29.78 ± 0.64 |
|
7 |
Cotton seed cake |
FH65 |
parasiticus |
260 ± 6.68 |
27.64 ± 0.32 |
|
8 |
Poultry Feed |
FH72 |
Flavus |
230 ± 5.55 |
25.03 ± 0.48 |
|
9 |
Poultry Feed |
FH74 |
flavus |
190 ± 4.44 |
23.17 ± 0.86 |
size of each primer pair produced a single DNA fragment is 1,032, 1,024 and 895 bp for apa-2, omt-1 and ver-1, respectively. Altogether, 102 samples were screened in which 09 (8.8%) samples were found positive to three targeted genes. Among 50 A. flavus, 06 (12%) were found positive. However, in 06 A. parasiticus 03 (50%) were positive to targeted genes. Furthermore, none of the A. niger species were found positive to these target genes. The amplified products were separated by Agarose gel electrophoresis as shown in Figure 2. The results are tabulated in Table 2.
Aflatoxin Detection by Thin Layer Chromatography
The results demonstrated the occurrence of only aflatoxin B1 and B2 in different food and feed samples contaminated by A. parasiticus and A. flavus. However, aflatoxin G1 and G2 were not found in any tested sample. In this study, 09 food and feed samples were investigated for the presence of aflatoxins. The obtained results confirmed that A. parasiticus and A. flavus have the ability to produce aflatoxins B1 and B2. FH44 strain of A. parasiticus was selected for further experiments as this strain produced highest amount of aflatoxins compared to other aflatoxigenic strains. The results are tabulated in Table 3.
Discussion
Mycotoxins are toxic compounds produced by fungi that are injurious to both humans and animals. These compounds contaminate various agricultural commodities like maize, cereals, spices, wheat, rice and animal feeds before, during and after harvest. Among them, aflatoxins are the widely studied and commonly known mycotoxins present in food and feed commodities. These toxins are produced as secondary metabolites by A. flavus and A. parasiticus in a range of food products.
In this study, 102 food and feed samples including composite spices, cotton-seed cake, maize, poultry feed, red chillies, rice and wheat were collected from different locations in Karachi-Pakistan. These samples were examined to check the total yeast and mould count (TYMC). All the tested samples were found contaminated at different levels having the highest average TYMC in cotton seed cake samples followed by poultry feed and maize samples. Altogether, the average count of all commodities was higher than 104 cfu/g. Fungal growth in food and feed samples is usually influenced by various factors. Climatic conditions, especially temperature and higher moisture content of the food samples play an important role in this process because the samples were collected in summer season and moisture content is normally high during that period. Moreover, it represents the unhygienic conditions of local market, improper handling and storage of samples. Similarly, most of the food items, crops, etc. are transported in uncleaned open goods carriers and trucks (Wagacha and Muthomi, 2008). According to Abbas (2005), high amount of moisture allows the growth of fungal species. Al-Hmoud et al. (2012) demonstrated the incidence of fungi in the food and feed samples and showed a wide variation that was in the range of 0.2x102 − 2.4x104 cfu/g. Similarly, Egbuta et al. (2015) reported a range of filamentous fungi including Aspergillus, Penicillium, Fusarium, etc., contaminating the food commodities. On one hand, these fungi deteriorate the quality of food products and on the other hand, they are capable of producing harmful toxins. It was also demonstrated that the Aspergillus species such as Aspergillus flavus, Aspergillus parasiticus were the most common toxigenic species in various grains, legumes, oil seeds, food and feed samples (Magnoli et al., 2002; Bueno et al., 2004; Egbuta et al., 2015).
During the current study, one hundred and two (102) strains of Aspergillus species were primarily isolated from the tested samples. These strains were identified as Aspergillus flavus (45.0 %), Aspergillus parasiticus (5.8%) and Aspergillus niger (49.0%) respectively. Aspergillus species, as reported earlier in several studies, were found to be the predominant fungal species in food and feed samples (Reddy et al., 2004). Atehnkeng et al. (2008) demonstrated the presence of Aspergillus flavus as predominant species among various Aspergillus in maize samples collected during a survey in Nigeria. Several similar studies have reported that the most prevalent fungi in pre and post-storage were Aspergillus (mostly A. flavus), Fusarium and Penicillium and their counts increased with longer storage period (Youssef et al., 2008; Azarakhsh et al., 2011). It was also demonstrated that the high prevalence of A. flavus is largely depends on long time storage in inappropriate conditions and unhygienic preparation. A. flavus possess a higher adaptability to growth substrates in a wide range of environment and the production of spores that remain viable even under extremely strict conditions (Saleemullah et al., 2006).
In this study, PCR was performed for the detection of aflatoxigenic genes i.e. ver-1, omt-1 and apa-2, respectively. These genes encode the key enzymes and regulatory factors in AFs biosynthesis pathway. A number of studies have demonstrated the significance of PCR based techniques to detect aflatoxigenic potential of Aspergillus strains (Shapira et al., 1996; Chen et al., 2002; Mayer et al., 2003). DNA extracted from the mycelia of three Aspergillus species was used as PCR template for each of the primer pairs. All samples were screened, out of which nine (09) samples were found positive to three targeted genes. In case of A. flavus, 06 Out of 50 strains were found positive for these genes, while 03 out of 06 of A. parasiticus isolates carried these genes. Similarly, previous study confirmed that the five Aspergillus species, four Penicillium species, and two Fusarium species were tested and among these, A. parasiticus and A. flavus were found positive for these genes (Shapira et al., 1996). Another study demonstrated the multiplex PCR with four sets of primers i.e. aflR, omt-A, ver-1 and nor-1 genes in which the clear detection of aflatoxigenic strains of A. flavus and A. parasiticus were observed (Criseo et al., 2001). However, none of the isolates of A. niger were found positive. It was reported that A. niger produces Ochratoxin A, but not aflatoxins (Schuster et al., 2002). Al-Hmoud et al. (2012) has demonstrated the prevalence of aflatoxinogenic A. parasiticus through PCR in food and feed samples in Jordan. In addition, real-time PCR (RT-PCR) method directed against the nor-1 gene of the aflatoxin biosynthetic pathway as target sequence was also used to scan Aspergillus flavus population in peanuts (Passone et al., 2010).
PCR results were further confirmed by thin layer chromatography (TLC). TLC was performed to confirm the presence of aflatoxin synthesis by Aspergillus species. All PCR positive strains were found positive to AFB1 and AFB2 as well, whereas, AFG1 and AFG2 were not detected in any tested sample. All nine positive strains have shown to produce high concentrations of total aflatoxins ranging from 220µg/kg to 552µg/kg. These concentrations are too high when compared to the maximum tolerable limits set by several regulatory authorities including 4µg/kg by European Union and 20µg/kg by United States Food and Drug Administration (USFDA) and Pakistan Standard and Quality Control Authority (PSQCA) (Asghar et al., 2014). Different concentrations of AFB1 and AFB2 were produced by A. flavus and A. parasiticus. FH44 strain of A. parasiticus produced the highest AFs amount in comparison to other strains. Several studies have reported that A. flavus usually produces AFB1 and AFB2 while A. parasiticus produces AFB1 and AFB2 as well as AFG1 and AFG2 (Al-Hmoud et al., 2012; Mishra and Das, 2003; Criseo et al., 2001; Ghadeer and Al-Delamiy, 2012). However, in this study, aflatoxigenic strains of A. parasiticus were not shown to produce AFG1 and AFG2. This study compared the detection of aflatoxin potential by two different methods i.e. PCR and TLC. PCR is rapid, less laborious, cost effective and highly sensitive method for the detection of particular genes involved in aflatoxin biosynthesis pathway. However, this technique cannot confirm that the said organism will produce aflatoxin. It might be possible that fungal strain does not produce AFs due to some internal and external factors. Sometime due to the stress, survival phase and mutation the organisms have lost its ability to produced AFs. Several studies have been demonstrated that the lack of aflatoxin production might be due to mutations including substitution of some bases and also many others different physiological conditions affecting aflatoxin biosynthesis (Erami et al., 2007; Liu and Chu, 1998). In this study, PCR methodology is used as a screening test for initial detection due to its speed and high sensitivity. The PCR positive strains of Aspergillus were examined for further investigation through TLC.
In comparison, TLC is also a simple and cost effective procedure to identify the AFs. It has an ability to directly detect the AFs production. However, there are some disadvantages related to this method such as, long incubation time, laborious extraction methods, insufficient sample clean-up resulting in poor separation, lack of precision, poor sensitivity and reduced resolution. It is more suitable for laboratories receiving only a few samples. Several studies previously reported the disadvantages of TLC (Gilbert and Anklam, 2002; Trucksess, 2005). Still, there are many organizations that are currently using TLC technique for aflatoxin detection on routine basis (Asghar et al., 2014; Bakhiet and Musa, 2011). In summary, it would be a better idea to use PCR for preliminary screening and subsequent confirmation of PCR positive strains by TLC for aflatoxin production.
On the basis of achieved results, it could be concluded that Aspergillus species are the most common fungal species present in food and feed products. In addition, aflatoxin producing strains, especially A. flavus and A. parasiticus are frequent in the food and feed products available in the local markets of Karachi, Pakistan. Hence, it is important to design and employ well-structured strategies to control these aflatoxigenic strains at pre- and post-harvest levels.
Acknowledgments
The authors are grateful to the department of Microbiology, Jinnah University for Women, Karachi and Food and Feed Safety Laboratory, PCSIR Laboratories Complex, Karachi for their full time support during this project.
conflict of interest
The authors declare no conflict of interest regarding this study.
authors’ contribution
Ibrahim F and Jalal H conducted all the experiments stated in this study. They also contributed in the writing of this manuscript. Khan AB, Asghar MA, Iqbal J designed this study and supervised all the experiments stated in this study. They also contributed in writing and finalizing this manuscript. Ahmed A and Nadeem G critically analyzed the data, contributed in writing and made the final decision for submission of this manuscript. All the authors approved the final version of this manuscript.
References






