Journal of Infection and Molecular Biology
Research Article
Application of Polymerase Chain Reaction Assay for Early Detection of Cell Free DNA (cfDNA) in Mice Infected with the Migratory Larvae of Trichinella spiralis
Rasha Abd Elmonem Hassan Attia1, Abeer Elsayed Mahmoud1, Ragaa Mohamed Othman1, Ayat Abd Elrahman Sayed2
1Departments of Parasitology; 2Medical Biochemistry, Faculty of Medicine, Assiut University, Egypt.
Abstract | To initiate effective treatment and to prevent fatal complication, diagnosis of human trichinellosis at an early stage is essential. Polymerase chain reaction (PCR) can be applied for early diagnosis of infection through amplification of Trichinella spiralis migratory larval DNA or their products from blood of infected mice. We evaluated the use of two PCR procedures for the detection of larval DNA from T. spiralis. Blood and plasma samples were collected from mice infected with 200 larvae of T. spiralis on days 4, 6, 14, 17 and 22 post infection (pi). PCR procedure with DNA extraction and direct PCR without DNA extraction were applied to amplify the target gene fragment (mitochondrial ATP6 synthase F0 subunit). PCR procedure with DNA extraction did detect T. spiralis migratory larval DNA in blood from days 4, 6, and 14 pi. PCR procedure without DNA extraction failed to amplify T. spiralis cell free DNA (cfDNA) from blood and plasma samples of infected mice. PCR assay with DNA extraction using ATP6 primers is a valuable procedure to detect T. spiralis migratory larval DNA in the blood of infected mice as early as day 4 and up to day 14 pi. Further validation of such assays on clinical samples would propose a promising diagnostic tool for the timely diagnosis of human trichinellosis.
Keywords | PCR, Cell free DNA, T. spiralis, Migratory larvae, Blood, Plasma
Editor | Tahir Yaqub, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | July 16, 2016; Accepted | July 27, 2016; Published | August 03, 2016
*Correspondence | Rasha AH Attia, Department of Parasitology, Faculty of Medicine, Assiut University, Assiut, Egypt; E-mail: rashaattia@gmail.com
Citation | Attia RAH, Mahmoud AE, Othnman RM, Sayed AA (2016). Application of polymerase chain reaction assay for early detection of cell free DNA (cfDNA) in mice infected with the migratory larvae of Trichinella spiralis. J. Inf. Mol. Biol. 4(1): 9-15.
DOI | http://dx.doi.org/10.14737/journal.jimb/2016/4.1.9.15
ISSN (Online) | 2307-5465; ISSN (Print) | 2307-5716
Copyright © 2016 Attia et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Trichinellosis is an important food-borne zoonotic disease worldwide (Wang et al., 2013). It carries public health risks and has an economic impact in porcine animal production and thus on food safety (Tang et al., 2015). It is caused by ingesting raw or poorly cooked meat containing larvae of the genus T. spiralis which is considered as the most pathogenic and cosmopolitan species that causes human trichinellosis (Tantrawatpan et al., 2013; Cui et al., 2015; Attia et al., 2015). In Egypt, T. spiralis is present in fresh and processed pork. Outbreak of human trichinellosis was documented in French tourists following the consumption of pork meat in 1975 with a prevalence rate of 4.5% in domestic pigs slaughtered at the Cairo Abattoir during the outbreak. Prevalence rate dropped to 1.7% in 1995–1999. However, risk of infection can still be considered. High prevalence of T. spiralis infection (13.3%) has been detected in rats from Alexandria abattoirs, in stray dogs, and in domestic pigs (Pozio, 2007; Youssef and Uga, 2014).
Early clinical diagnosis of trichinellosis in man is difficult because of the varied and untypical symptoms. Its diagnosis during early stages of infection is essential to start an effective treatment and avoid deadly complications (Watt et al., 2000; Dupouy-Camet et al., 2002; Machnicka et al., 2001). The techniques that are used (immunoradiometric assay or monoclonal antibodies sandwich ELISA) are not applicable for the serological diagnosis of trichinellosis and give false-negative results during the early stages of infection in human and experimentally infected mice (Dupouy-Camet et al., 2002; Wang et al., 2007; Li et al., 2010). Detection of circulating larvae or their products in blood by PCR is useful for early diagnosis of trichinellosis (Pozio and Darwin, 2006; Golab et al., 2009; Li et al., 2010). A major problem with PCR-based diagnostic tests of blood samples is the false-negative or low-sensitivity reactions caused by PCR inhibitors despite the advanced DNA purification methods (Zhang et al., 2010).
The detection of circulating cell-free DNA (cfDNA) in body tissues or fluids including human plasma has been recognized for the non-invasive diagnostic purposes of a diversity of clinical conditions in medicine (Lo and Chiu, 2007; Goessl, 2003) including oncology and prenatal diseases (Chen et al., 1996; Sozzi et al., 2003; Zimmermann et al., 2007; Chiu and Lo, 2004; Lo and Poon, 2003). In parasitic diseases, there is a huge turnover of parasites involving replication, maturation, migration, friction during penetration and death which liberate DNA into the blood. Using PCR for the early detection of free circulating DNA (cfDNA) in blood and plasma was assessed in certain parasites as Toxoplasma gondii (Weiss et al., 1991), Trypanosoma cruzi (Kirchhoff et al., 1996) and schistosomiasis (Hussein et al., 2012; Wichmann et al., 2009).
The present procedures for the detection of cfDNA apply DNA extraction and purification which is time-consuming and has many limitations that might shift the DNA integrity values (Loparev et al., 1991; Clausen et al., 2007; Huggett et al., 2008; Fong et al., 2009; Fleischhacker et al., 2011). To overcome this, an approach is used for the detection of cfDNA directly in plasma or serum without preceding DNA extraction (Schwarzenbach et al., 2011; Umetani et al., 2006). We assume that it might be possible to detect the circulating cell-free DNA (cfDNA) of T. spiralis migratory larvae in the blood or plasma of experimentally infected mice as well.
The current study aimed to test the use of PCR with and without DNA extraction for the early detection of T. spiralis migratory larval cfDNA in the blood and plasma of experimentally infected mice.
Material and Methods
Experimental Design
In the current study, we used a strain of T. spiralis which was isolated originally from naturally infected pigs obtained from El-Bassatine Abattoir, Cairo. It was maintained by routine serial passages in BALB/c mice at the animal facility, Faculty of Medicine, Assiut University, Assiut, Egypt. The mice were infected following the method described previously (Gamble, 1996). The collection of larvae for mice infection in this study was done as follows: The infected mice were killed, skinned and eviscerated on day 40 pi. The carcasses were minced and digested for 4 hours at 43°C in a solution of 0.33% pepsin and 1% HCl. The whole digest was poured and settled for 30 minutes. About 40mL of the fluid is drained from the fluid into a 50mL glass centrifuge tube. It was settled for another 10 minutes, and the upper fluid is aspirated and discarded. The last 10 mL of sediment is poured into a petri dish and examined for larvae (Gamble et al., 2000). To determine the number of larvae/ml, we used a micropipette and placed 10 0.05 ml drops on a petri dish and counted the total amount of larvae with a microscope. The procedure was performed in duplicate (Goettsch et al., 1994).
A total of sixty pathogen free BALB/c mice (25-30 g) aged six to eight weeks were purchased from the animal facility, Faculty of Medicine, Assiut University, Assiut, Egypt, and maintained under controlled light and temperature with standard diet and water supplies. Fifty of them were orally infected with about 200 muscle larvae of T. spiralis per mouse while the remaining 10 were kept as the uninfected control group. The fifty mice were divided into five groups (10 mice each). Each group of mice was used in one of these selected days, i.e. 4, 6, 14, 17, and 22 post infection (pi). Blood samples of mice of one group were collected at the five selected days (days 4, 6, 14, 17, 22 pi). About 1 ml of blood was collected from each mouse via retro-orbital puncture (Franssen et al., 2011) and plasma was separated by centrifugation at 1500 x g for 10 min. The collected amount of blood and plasma from each mouse was divided into 2 halves; half the amount was used for PCR with DNA extraction while the other half was used for PCR without DNA extraction. Both blood and plasma samples were stored at -20°C until used for PCR. Mice of all groups were sacrificed immediately after the collection of blood samples on each day.
Polymerase Chain Reaction (PCR)
DNA was extracted from 200 µl of whole blood and plasma collected from infected mice using Qiagen mini spin column according to the manufacturer’s instruction. DNA was also extracted from isolated muscular larvae and eluted so that DNA equivalent to 5 larvae was re-suspended in 1 µl DNase free water. Serial dilutions of larval DNA in DNase free water were prepared to have 10, 5, 1, 1/10, 1/20, 1/50, and 1/100 larval DNA/10 µl. In the meantime blood from non-infected mice was spiked with larval DNA to get the same serial dilutions but in blood and plasma.
The following pair of primers, ATP6-F 5′-CACACTAACCAAAGCCAAACCATC-3′ and ATP6-R 5′-GGAGTATGTTAGATGTTATTGTGTAGGAG-3′ (Golab et al., 2009), was used to amplify 250 nucleotides of the ATP6 ATP synthase F0 subunit gene (GeneID: GU33913). PCR was carried out using Thermo Scientific Phusion Blood Direct PCR Kit (Cat no. F547S, Thermo ScientificTM) designed to amplify both genomic and extra genomic target DNA by direct PCR from blood and body fluids including plasma. It contains Phusion Hot Start II High-Fidelity DNA Polymerase characterized by high resistance to PCR inhibitors found in blood even with samples containing 40% whole blood. First, the performance of the system was examined using the 10 µl of larval DNA serial dilution; both in DNase-free water and in blood. The sample volume represented 20% of the total reaction volume, in which 25 µl of PCR master mix, 1UM of each primer, and DNase-free water to final reaction volume of 50 µl. PCR conditions were as follows: denaturation at 95 °C for 10 min, 35 cycles at 95 °C for 30 sec, 55 °C for 30 sec and 72 °C for 1 min, followed by an extra extension step at 72 °C for 10 min. After validation of the method, PCR was carried out using 10 µl of blood, plasma, or DNA extracted from blood and plasma of infected mice. In each PCR a positive control sample representing DNA equivalent to single larva and a non-template control (NTC) were included. PCR products were visualized on 1% ethidium bromide stained agarose gel and visualized by Biodoc-It gel documentation system.
Ethics
The experimental animal studies were conducted in accordance with the international valid guidelines and were maintained under convenient conditions at the Animal House, Faculty of Medicine, Assiut University, Egypt.
Results
PCR Amplification of T. spiralis Larvae Gene
PCR amplification of the muscle larval DNA corresponding to one larva, 1/10, 5 and 10 larva using the ATP6 primer yielded a 250bp DNA fragment. The same pattern was obtained when the amplification was carried out from 50 µl of uninfected mouse blood or plasma spiked with the same amount of larval DNA (Figure 1).
PCR Detection of Blood and Plasma of Mice Infected with T. spiralis Larvae
PCR amplification of extracted DNA from blood samples of infected mice yielded T. spiralis migratory larval DNA at 250bp fragment only on days 4, 6 and 14 pi (Figure 2A) while no yield was detected on days 17 and 22 pi. When the PCR procedure with DNA extraction was performed on the 10 blood samples from 10 mice of each group on days 4, 6 and 14 pi, the DNA was amplified from five samples on each selected day (50% detection rate). However, the same PCR procedure did not detect the extracted larval DNA in plasma samples taken from mice on the previously mentioned days.
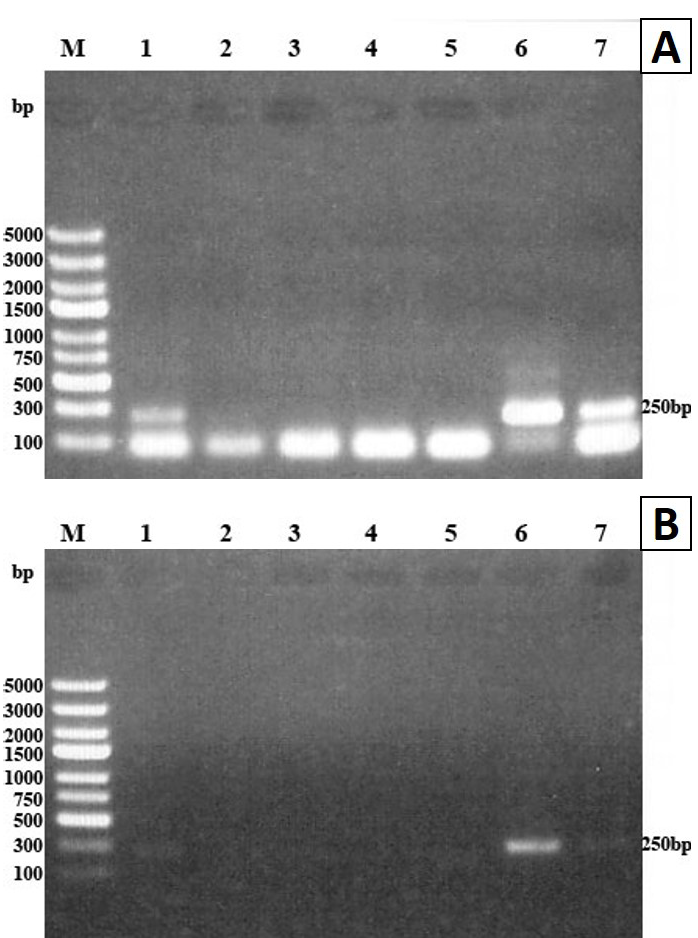
Figure 1: PCR of DNA extracted from T. spiralis muscular larvae using ATP6 primer. PCR products were analyzed by 1% agarose gel and stained with ethidium bromide
A) Amplification from one larva, 1/10, 1/20, 1/50, 1/100, 5 larva and 10 larvae (lanes 1-7 respectively), lane M molecular weight marker; B) Amplification from the same larval DNA spiked with 50 µl blood of uninfected mouse. PCR products were detected in lanes 1, 2, 6 and 7 at 250bp DNA fragment
The PCR procedure without DNA extraction failed to amplify cfDNA of T. spiralis migratory larvae in blood and plasma of infected mice on any of the selected days, i.e. days 4, 6, 14, 17 and 22 pi (Figure 2B).
Discussion
Trichinellosis involves a wide range of symptoms and its accurate diagnosis in humans is difficult. During early Trichinella infection, there is continuous production of newborn larvae which enter and circulate into the blood until the expulsion of adult worms from the intestine. Direct microscopic observation of these larvae in the blood is too laborious (Li et al., 2010).
In this study, PCR after DNA extraction and direct PCR without DNA extraction of blood and plasma samples were used to detect T. spiralis migratory larval DNA in infected mice on the selected days, i.e. days 4, 6, 14, 17, 22 pi. Our results showed that PCR after DNA extraction detected T. spiralis migratory larval DNA from blood samples on days 4,6,14 pi. This result showed that the PCR procedure with DNA extraction is valuable for early detection of T. spiralis migratory larval DNA from blood samples of experimentally infected mice. In the current study, PCR detection of Trichinella DNA in blood and plasma samples started from day 4 up to day 14 pi, which corresponds to the period in which larval deposition by adult females begins, and lasts for the next 10–20 days while the new-born larvae migrate through blood. Consistent with our results, the PCR procedure detected circulating T. spiralis larvae in the blood of mice from day 5 to day 14 pi (Uparanukraw and Morakote, 1997; Li et al., 2010) and from day 5 until day 17 pi (Caballero-Garcia and Jimenez-Cardoso, 2001). However, we detected Trichinella DNA earlier on 4 day pi. This difference between the results may be due to the discrepancy in DNA extraction procedures from blood samples and/or to different parasitic burdens (Li et al., 2010).
(A) and plasma (B) on days 4, 6, 14 pi using ATP6 primers. PCR products analyzed by 1% agarose gel and stained with ethidium bromide. Lane M molecular weight marker, lanes 1, 2 and 3 blood samples from days 4, 6 and 14 pi, lane 4 positive control (DNA extracted from one larva), lane 5 non-template control (NTC). The amplified DNA fragment was detected at 250bp in lanes 1, 2, 3 and 4
The detection rate of the PCR from blood samples of infected mice was about 50% of the infected mice as 5 of 10 blood samples from 10 mice of each selected day were positive by PCR on days 4, 6, and 14 pi. The variable results of the PCR procedure on the same day are perhaps due to discrepancies in the extraction of blood samples in the same or different experiments. Also, the efficacy of DNA extraction could explain the varied results produced from the blood samples containing small numbers of migratory larvae (Uparanukraw and Morakote, 1997).
A main problem with PCR-based diagnostic procedures of blood specimens is the false-negative or reduced-sensitivity reactions produced by PCR inhibitors despite the advanced DNA purification procedures used. In spite that, serum and plasma specimens contain fewer PCR inhibitors than whole blood specimens (Zhang et al., 2010), our results showed that PCR with DNA extraction from plasma samples failed to detect T. spiralis cfDNA on any day during the whole experiment. This may be due to the fact that the detection rate of some pathogens in serum and plasma specimens can be lesser than that from whole blood specimens as some pathogen fractions are maintained in the peripheral cells (Watkins-Riedel et al., 2004; Klungthong et al., 2007).
The concept of using circulating cell-free DNA in human plasma for diagnosis has long been discovered and proven in oncology and prenatal diseases (Chen et al., 1996; Sozzi et al., 2003; Zimmermann et al., 2007; Chiu and Lo, 2004; Lo and Poon, 2003). In parasitic diseases such as schistosomiasis, PCR for detection of cell free DNA in human plasma might carry the potential of a new laboratory method for diagnosing any stage of clinical schistosomiasis (Wichmann et al., 2009). The detection of free circulating DNA by PCR can be a valued test for the early diagnosis of prepatent S. mansoni infection in mice (Hussein et al., 2012). Similarly with other parasites, Toxoplasma gondii and Trypanosoma cruzi DNA was detected by PCR during early infection stages in the experimental animals, thus permitting the diagnosis of infection earlier than when using conventional diagnostic procedures such as microscopy or serology (Weiss et al., 1991; Kirchhoff et al., 1996).
So, our hypothesis was that the detection of cell free T. spiralis migratory larval DNA in the blood of mice may become a realistic choice to diagnose the early stage of infection. The current procedures for qualitative cell free DNA analysis imply DNA purification. Unfortunately, DNA purification methods have been proven to be time-consuming and susceptible to mistakes or DNA losses (Huggett et al., 2008; Brojer et al., 2005; Fleischhacker et al., 2011). So, we reasoned that the direct PCR procedure without DNA extraction may be more sensitive for the detection of cell free DNA (cfDNA) from blood or plasma. Yet, the use of PCR of blood or plasma without DNA extraction in our study failed to detect T. spiralis migratory larval cfDNA on any day during the whole experiment.
To our knowledge, this is the first attempt to use direct PCR without DNA extraction for the detection of T. spiralis migratory larval cfDNA from the blood and plasma of mice. Unfortunately, this attempt yielded no amplification of T. spiralis larval DNA, which may be due to low sensitivity of the method used in the study, or to the absence of parasitical DNA in the examined samples (Golab et al., 2009). We assumed that one possible explanation is the need to use more than 20% of the PCR final reaction volume to increase the amount of DNA in the examined samples and reliability for its detection, but this is not recommended as by increasing the blood amount, the PCR inhibitors in the blood will increase as well and lower the sensitivity of the procedure.
conclusion
The results of the current study showed that the PCR technique using a suitable primer (ATP6) can be useful for the early detection of T. spiralis migratory larvae in blood of experimentally infected mice even from day 4 pi. Therefore, this technique may be an effective tool for the early diagnosis of trichinellosis in humans as well, especially those in whom serology (ELISA test) for specific anti-Trichinella antibodies yielded negative results. Unfortunately the use of the direct PCR procedure for the detection of T. spiralis migratory larvae cfDNA from the blood or plasma of mice without previous DNA extraction showed negative results. Further trials are needed to implement this procedure with optimization between the uses of different PCR reaction volumes and / or increase the amount of the blood used.
Acknowledgments
Our special acknowledgment to Dr. Ashref Girgis Abd Elmalak, lecturer of english language, Faulty of Arts, Assuit University who helped us revising the English version of this manuscript.
Conflict of Interest
The authors don’t have any conflict of interest to declare concerning this article.
Authors’ Contributions
Rasha A.H. Attia, Abeer E. Mahmoud, Ragaa M. Othnman conceived the idea, designed and carried out the experimental work. Ayat A Sayed performed the PCR procedure. All authors helped in data analysis, collection of papers, writing and revising the manuscript before submission.
References






